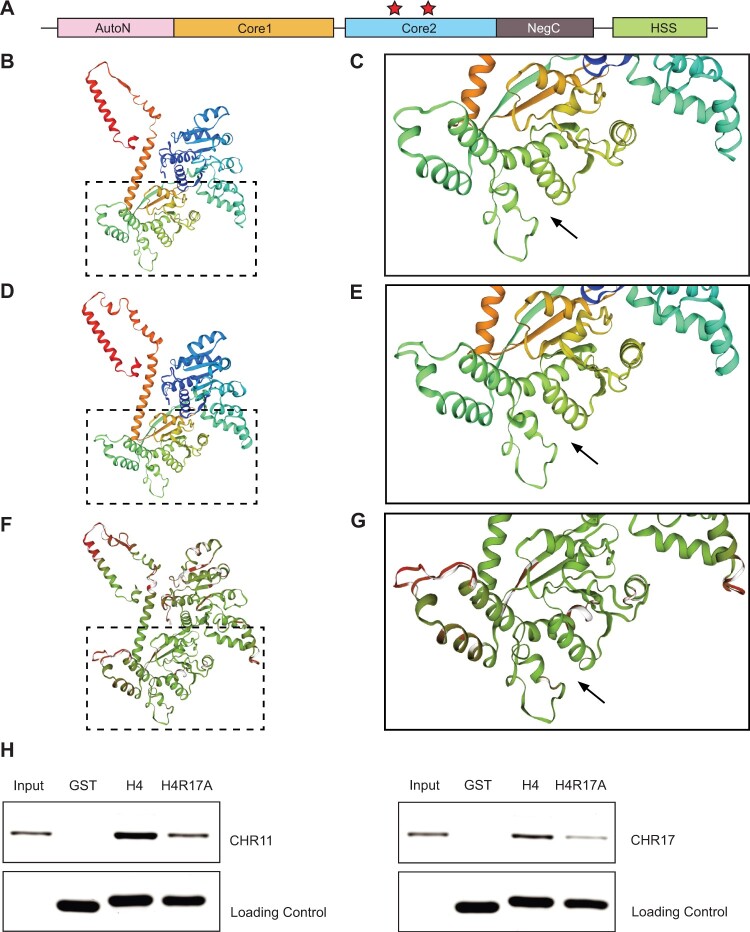
Figure 4.
The H4R17A mutation prevents binding by Arabidopsis ISWI enzymes. A, Domain architecture of ISWI. HSS, HAND–SAND–SLIDE; core1, first RecA-like domain; core2, second RecA-like domain. Red stars indicate the residues implicated in binding H4R17 on the second RecA-like ATPase core domain (core2) identified in M. thermophila (Yan et al., 2016) and S. cerevisiae (Yan et al., 2019). B and C, Homology model of Arabidopsis CHR11 amino acids (aa) 176–706. D and E, Reference structure of M. thermophila ISWI (5JXR) aa 173–718. F and G, Superposition of Arabidopsis CHR11 and M. thermophila ISWI structures with consistency color scheme (green indicates more consistent and red indicates less consistent). Black arrow denotes the predicted (Arabidopsis) or validated (M. thermophila) binding pocket of histone H4 arginine 17 (Yan et al., 2016). The boxed regions are enlarged in (C), (E), and (G). H, In vitro pull-down assay with recombinant GST or GST-tagged histone proteins (H4 or H4R17A) and CHR11 (left) or CHR17 (right).