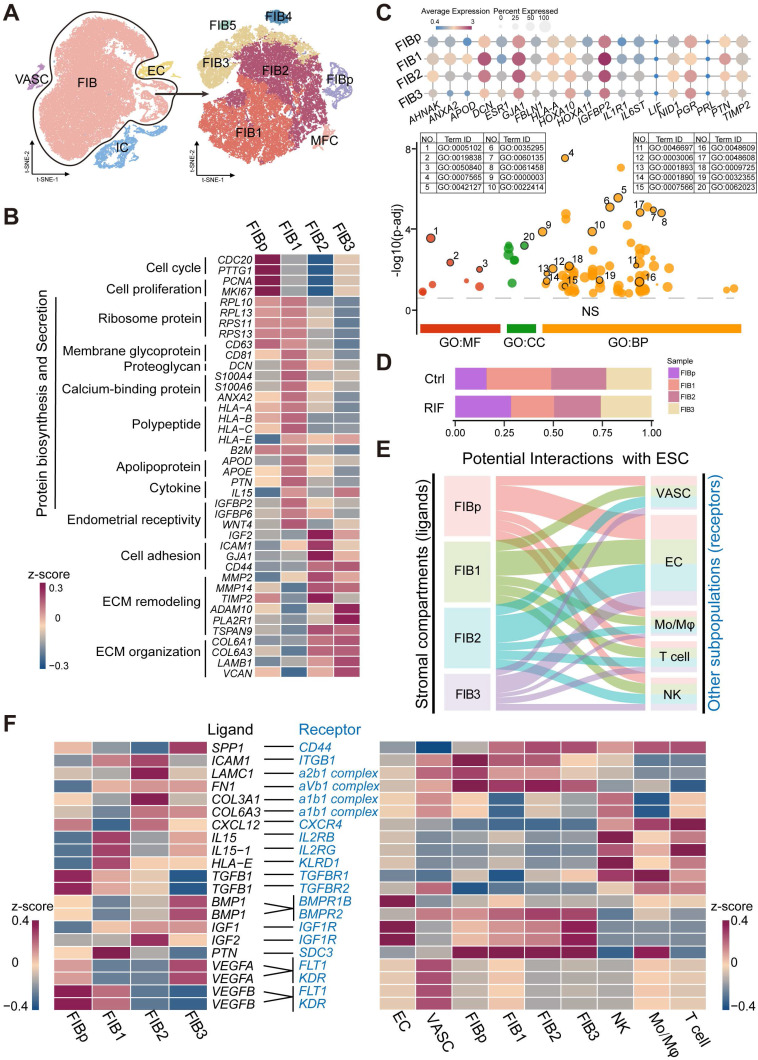
Figure 2.
Identification of population of human endometrial FIB at the WOI. (A) Cell cluster of FIB (52825 cells) was re-clustered into seven sub-clusters visualized by t-SNE. (B) Heat map showing relative expression (z-score) of selected genes for four main sub-clusters of endometrial FIB. (C) Bubble diagram showing average expression of endometrial receptivity-related genes (such as HOXA10, IGFBP2, LIF, and PRL) for four FIBs (FIBp, FIB1, FIB2 and FIB3) (up). Manhattan plot indicating the Gene Ontology (GO) enrichment of endometrial receptivity-related genes, and the most important GO terms (the details of GO terms are showed in Figure S6) of endometrial receptivity-related genes (down). One dot represented one GO term, and the size of the dots represented the number of genes that were enriched. The Manhattan plot was drawn by an online tool - g:Profile (https://biit.cs.ut.ee/gprofiler/). (D) Proportions of the four FIBs between the Ctrl and RIF groups. (E) Potential interactions between four FIBs and other subtypes of endometrium cells based on receptor-ligand pairs analysis. Width of the line represented the number of receptor-ligand pairs. (F) Heat map showing selected significant ligand-receptor interactions (P value < 0.05, permutation test, see Methods) between four FIBs (left) and endometrium cells (right). Assays were carried out at the mRNA level, but are extrapolated as protein interactions.