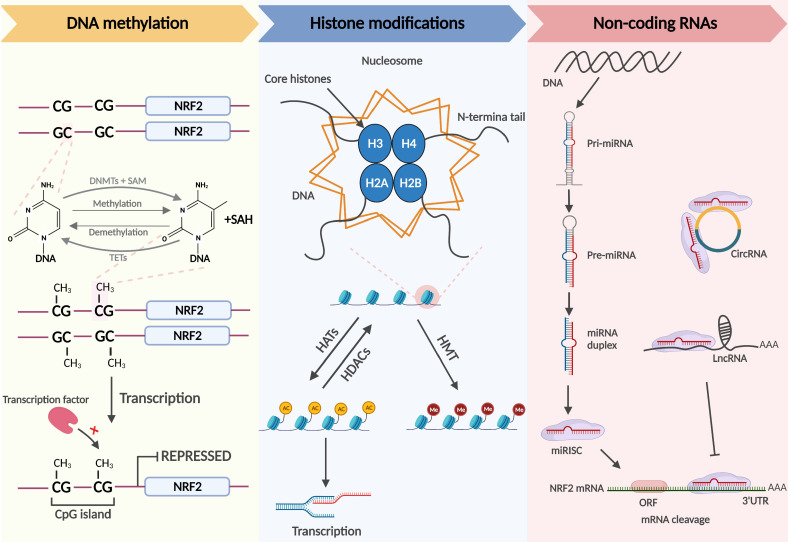
Figure 2.
Nrf2-related epigenetic modifications: DNA methylation. DNMTs methylate the C5 position of cytosine in DNA to generate 5-methylcytosine, which is progressively oxidized to 5-carboxylcytosine by TETs. When the CpG islands in the promoter region are extensively methylated, the transcription factor cannot identify the promoter, and Nrf2 transcription is repressed. Histone modifications. Chromatin consists of DNA and histone complexes. In each nucleosome, four core histone proteins (H2A, H2B, H3, H4) form an octamer around which DNA is tightly wrapped. Histone tails, the free amino ends of histones, can be modified differently under the action of related enzymes, affecting gene expression. Histone acetylation and deacetylation are regulated by HATs and HDACs and hyperacetylated histones are associated with transcriptional activation. Histone tail lysine and arginine residues are methylated by HMTs. The effect of histone methylation on transcription depends on the methylation site and the methylation stoichiometry. Noncoding RNAs. Mature miRNAs identify the complementary bases of the 3'UTR in the Nrf2 mRNA and trigger mRNA cleavage. CircRNAs and lncRNAs contain miRNA binding sites that act as competing endogenous RNAs (ceRNAs) to regulate the expression of miRNA target mRNAs.