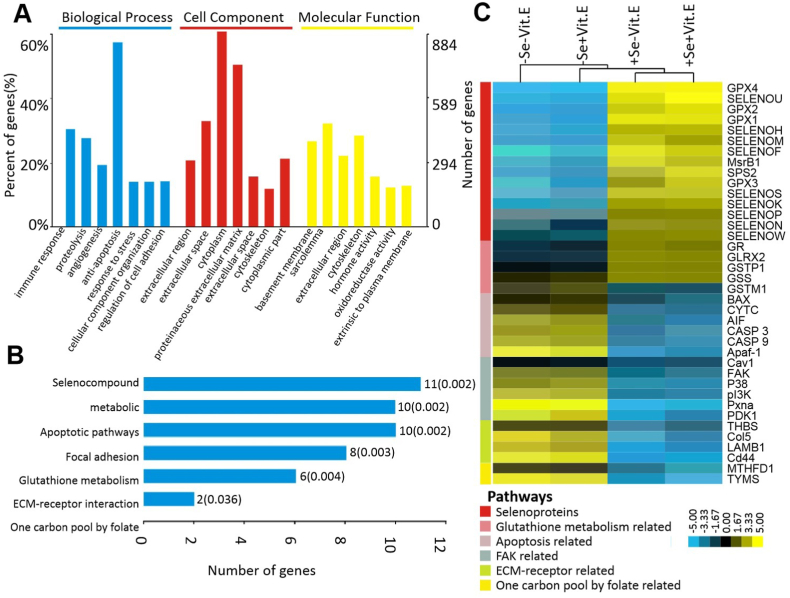
Fig. 2.
The GO analysis (A) with GO database and the pathway analysis (B) with KEGG database were applied for functional annotation. The significant GO terms and pathways were defined as P < 0.05 and FDR <0.05. (C) Heatmap showing the significant top 38 mRNAs affected by dietary Se and Vit. E deficiency in the pancreas of chicks on day 18. Yellow: increased; blue: decreased. P < 0.05 and FDR <0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Abbreviations: Se, selenium deficient; +Se, selenium adequate; –Vit. E, vitamin E deficient; +Vit. E, vitamin E adequate; Apaf-1, apoptotic peptidase activating factor 1; AIF, apoptosis inducing factor; BAX, BCL2 associated X; CASP 3, caspase 3; CASP 9, caspase 9; Cav1,caveolin 1; Cd44, CD44 molecule; CYTC, cytochrome c; Col 5, collage 5; GO, gene ontology; GPX1, glutathione peroxidase 1; GPX2, glutathione peroxidase 2; GPX3, glutathione peroxidase 3; GPX4, glutathione peroxidase 4; GLRX2, glutaredoxin 2; GR, glutathione reductase; GSS, glutathione synthetase; GSTM1, glutathione S-transferase mu 1; GSTP1, glutathione S-transferase pi 1; LAMB1, laminin subunit beta 1; KEGG, kyoto encyclopedia of genes and genomes; MsrB1, methionine sulfoxide reductase B1; MTHFD1, methylenetetrahydrofolate dehydrogenase, cyclohydrolase and for myltetrahydrofolate synthetase 1; p38, p38 kinase; PDK1, pyruvate dehydrogenase kinase 1; PI3K, phosphatidylinositol 3-kinase; Pxna, paxillin a; SELENOF, selenoprotein 15; SELENOK, selenoprotein K; SELENOH, selenoprotein H; SELENOM, selenoprotein M; SELENON, selenoprotein N; SELENOP, selenoprotein P; SELENOS, selenoprotein S; SELENOU, selenoprotein U; SELENOW, selenoprotein W; SPS2, selenophosphate synthetase 2; THBS, thrombospondin 1; TYMS, thymidylate synthetase.