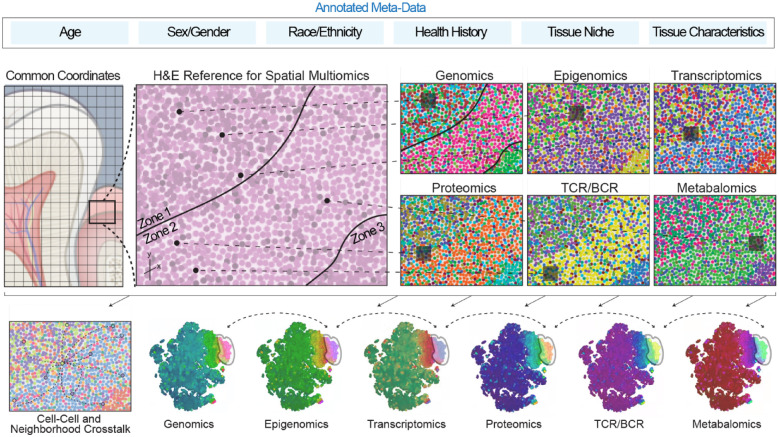
Figure 3.
Applied spatial multiomics for coordinated and integrated analyses. Coordinated efforts across the Human Cell Atlas are working toward building a consensus of the necessary metadata (clinical and biological) to generate a common coordinate framework for the human body. Future work including these additional layers of data will highlight the diversity of cell types and states across humans considering age, sex, race, ethnicity, ancestry, and oral and systemic health history, as well as the specific niche, tissue orientation, and health status of that niche (healthy, inflamed) at the time of sample collection. There is an immense need to interrogate a higher number of molecular dimensions or human tissues—genome, transcriptome, epigenetic modifications, proteome, T cell receptor and B cell receptor repertoires (TCR/BCR), and metabolome of the collected sample itself—as no single “omics” technology can fully define the complexity of molecular mechanisms, but taken together, these integrated data have the potential to provide a more comprehensive landscape of basic biological processes and human disease. Multimodal sequencing has the capacity to move the field from descriptive “snapshots” toward a mechanistic understanding of gene regulatory networks and, importantly, to refine sources of cellular heterogeneity as already applied to the immune system. The use of multimodal single cell and spatial multiomics is therefore revolutionizing our understanding of cellular biology; however, relying on the dissociation of cells from their natural tissue environment limits our ability to understand the role of intrinsic and extrinsic factors that underpin cellular communication and organ function. Spatial multiomic approaches, which include information on the location of cells, will still need to be integrated with these single cell multiomic maps.