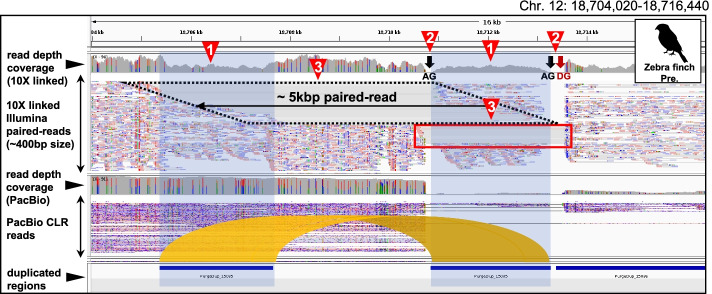
Fig. 3.
The presence of a gap and discordant reads between false duplications. Shown is a locus in the previous zebra finch assembly with a false duplication. 10X linked read alignments are shown above the PacBio CLR read alignments, along with the depth coverage of the respective read data. Characteristics of false duplications are marked with red triangles: (1) Nearly half depth-coverage and lack of heterozygous variants—colors indicating nucleotide heterozygosity; (2) gaps between false duplications; and (3) discordant 10X linked reads (black dotted box). Red box, discordant reads found near the end of scaffolds that should be connected to each other. AG, assembly gap. DG, depth-gap (unsupported sequences by reads; see the “ Materials and methods” section)