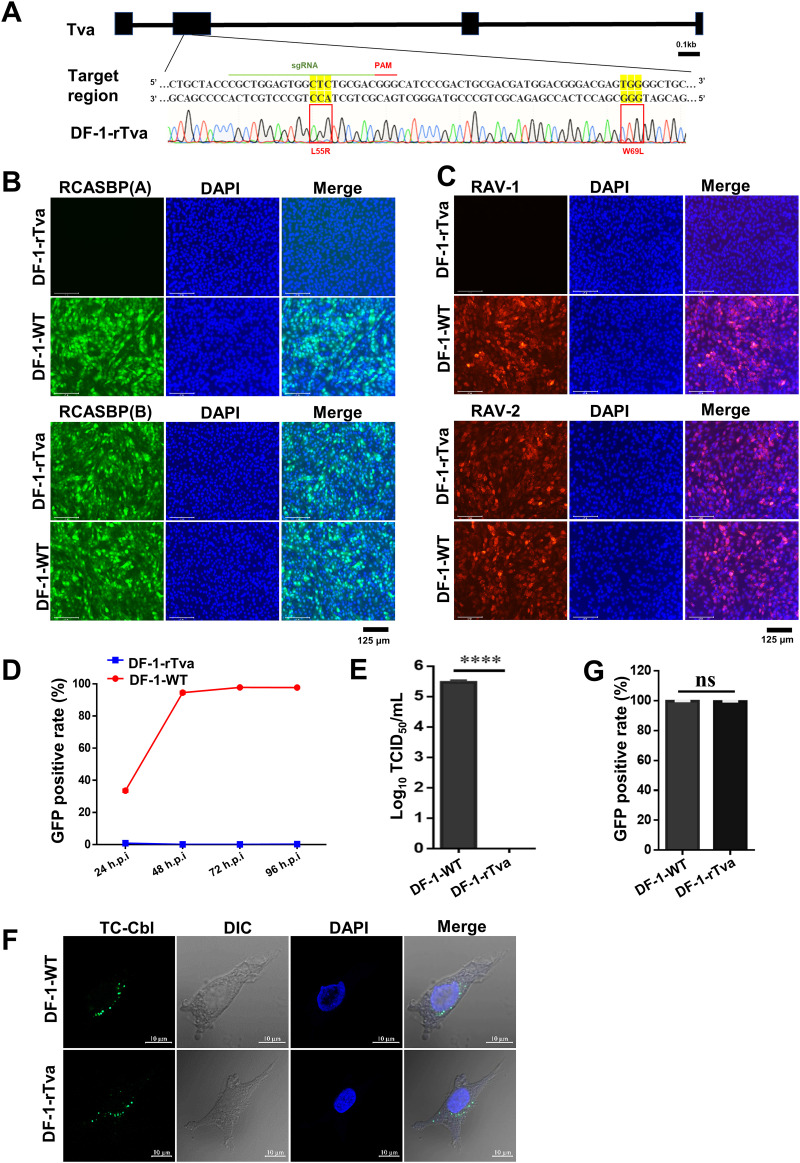
FIG 8.
Precise gene editing of both L55 and W69 of chicken Tva conferred resistance to ALV-A. (A) Schematic diagram of the construction of Tva gene-edited DF-1 cells and sequence analysis of the DF-1-rTva cells. (B) DF-1-WT and DF-1-rTva cells were challenged with RCASBP(A) or RCASBP(B) at an MOI of 1 and analyzed by fluorescence microscopy at 120 hpi. Scale bar: 125 μm. (C) DF-1-WT and DF-1-rTva cells were challenged with RAV-1 or RAV-2 (wild-type ALV-B strains) at an MOI of 1 and analyzed by fluorescence microscopy using an anti-ALV-A monoclonal antibody at 120 hpi. Scale bar: 125 μm. (D) DF-1-WT and DF-1-rTva cells were infected with RCASBP(A) at an MOI of 1 and analyzed by flow cytometry at 24, 48, 72, and 96 hpi. (E) Entry of RAV-1 virus into DF-1-WT or DF-1-rTva cells as analyzed by measuring virus titers in cell supernatants collected 7 dpi. (F) Confocal microscopy analysis of TC-Cbl uptake in DF-1-WT and DF-1-rTva cells. Nuclei were counterstained with DAPI. Scale bar: 2 μm. (G) TC-Cbl uptake in DF-1-WT and DF-1-rTva cells as analyzed by flow cytometry. Data from three independent experiments are shown as means ± standard deviations of triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significant difference.