Figure 3.
Remotely donated tumor and cell-free DNA samples obtained through patient partnership recapitulate known genomic findings in metastatic prostate cancer
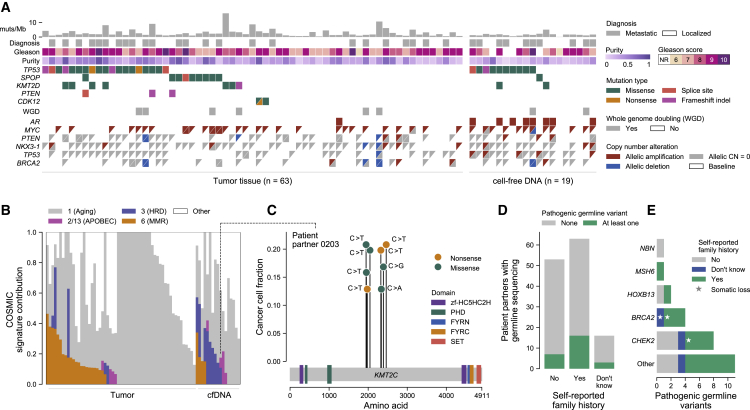
(A) Genomic and clinical landscape of 82 sequenced samples. Columns represent samples, separated into tumor (prostate, left) and cell-free DNA (cfDNA; donated blood, right) samples, while rows represent select clinical and genomic features. Gleason scores for tumor samples are taken from the pathology report received with the sample (n = 58) or the patient partner’s medical records (n = 5) if Gleason scores were not provided in the report. Gleason scores for cfDNA were taken from pathology reports in the medical record, with NR representing cases where a Gleason score was not reported in the medical record. Diagnosis refers to whether the initial diagnosis of prostate cancer was localized or metastatic. Multiple mutations in the same gene are represented as triangles. WGD refers to whole-genome doubling. Copy-number calls are allelic and defined with respect to baseline allelic ploidy (2 for samples with WGD, one for those without), with calls for the two alleles indicated by two triangles (except for AR, which has only one allele in men and so is shown as a single box). Allelic CN = 0 refers to complete allelic deletions. Allelic deletions that are not complete deletions are possible in samples with WGD. Figure created with CoMut.29
(B) Mutational signature analysis of sequenced samples. The relative contribution of select COSMIC v.2.0 mutational signatures are shown, separated by tumor and cfDNA (donated blood) sample type.30 APOBEC refers to signatures associated with activity of APOBEC family of cytidine deaminases (signatures 2 and 13); MMR to the signature associated with deficient DNA mismatch repair (signature 6); and HRD to the signature associated with homologous recombination deficiency (signature 3). To be denoted as present, a signature cutoff of 6% was used. Samples with too few mutations for signature analysis (<50 mutations, n = 5 samples) are not shown.
(C) Instance of localized hypermutation (kataegis) of KMT2C in cfDNA from a donated blood sample. The y axis shows the cancer cell fraction of each mutation, while the x axis shows their amino acid within KMT2C. Domains taken from Pfam.31 The dotted line connects to this sample’s mutational signature profile.
(D and E) Germline pathogenic alterations and their overlap with patient-reported family history. Pathogenic germline alterations (as annotated by ClinVar) in genes from a select panel of genes previously implicated in cancer heritability were detected in patient partners with sequenced saliva or blood buffy coat (n = 132) (STAR Methods; Tables S3 and S5).32 Survey responses to a question asking about a family history of prostate or breast cancer were tabulated and overlapped with this genomic data. Stars in (E) indicate instances where a somatic deletion also affected that gene in a tumor or cfDNA sample from that patient partner, suggesting biallelic inactivation.