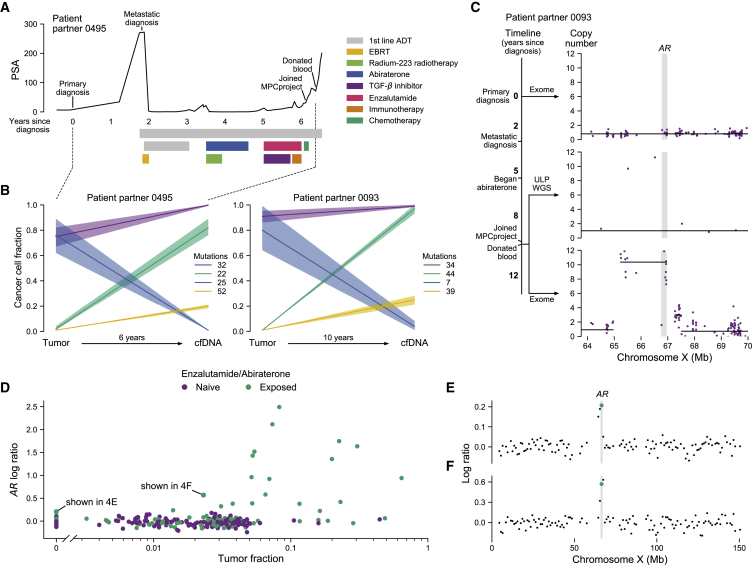
Figure 4.
cfDNA from donated blood reveals patterns of clonal dynamics and clinically relevant genomic changes
(A) Clinical trajectory of patient partner 0495. This patient partner’s prostate-specific antigen (PSA) trajectory is shown on the y axis, time in years since initial diagnosis is shown on the x axis, and bars denote the beginning and end of therapies. EBRT, external beam radiation therapy; first-line androgen deprivation therapy (ADT), leuprolide and bicalutamide; immunotherapy, nivolumab; chemotherapy, cisplatin and etoposide.
(B) Tumor evolution from primary tumor to metastatic cfDNA samples. The y axis shows the cancer cell fraction (CCF) of clonal clusters identified between tumor and cfDNA samples (x axis). Time between samples shown on the x axis. Colors indicate how many mutations were identified in each clone, with a 95% confidence interval around the estimated CCF. Purple represents the truncal/ancestral clone. Clusters with CCF <0.10 across all biopsies are omitted. The clinical trajectory of patient partner 0495 (left) is shown in (A), while the trajectory of patient partner 0093 (right) is shown in (C).
(C) Emergence of AR amplification in patient partner 0093 induced by anti-androgen therapy. The timeline depicts this patient’s clinical trajectory, while the plots show the absolute copy number (y axis) of the genomic region around AR (x axis, gene body shown in gray). The first plot depicts exome sequencing from the patient’s archival tumor tissue; the second and third plots depict ultra-low-pass whole-genome sequencing (ULP-WGS) and exome sequencing of cfDNA from the patient’s donated blood, respectively. Individual points represent copy number of target regions (exome) or copy number of 1 Mb genomic windows (ULP-WGS). Black lines represent discrete copy-number segments.
(D–F) ULP-WGS reveals clinically relevant AR amplifications even at low tumor fraction. In (D), tumor fraction of 318 cfDNA samples from donated blood of 300 patient partners with ULP-WGS sequencing is shown on the x axis, while the log copy ratio (logR) of the genomic interval containing AR is shown on the y axis. Points are colored by whether patient partners self-reported taking enzalutamide or abiraterone. 89 samples are shown with tumor fraction of 0 (undetectable), while 229 have non-zero tumor fractions. Two samples, one at a tumor fraction of 0 and another at a tumor fraction of 0.023, have chromosome X log copy ratio profiles shown in (E) and (F), respectively. The green points represent the values shown in (D), with the genomic interval containing AR highlighted in gray.