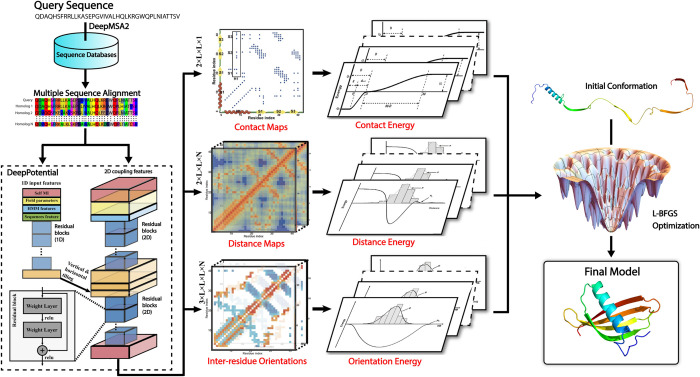
Fig 1. Overview of the DeepFold pipeline.
Starting from a query amino acid sequence, DeepMSA2 is used to search the query against multiple whole-genome and metagenome sequence databases to create a multiple sequence alignment (MSA). The MSA is then used by DeepPotential to derive input features based on co-evolutionary analyses for the deep ResNet training. DeepPotential outputs the probability distribution of the Cβ-Cβ/Cα-Cα contact and distance maps as well as the inter-residue orientations. These restraint potentials along with the inherent statistical energy function are used to guide the L-BFGS folding simulations for final full-length structure model construction.