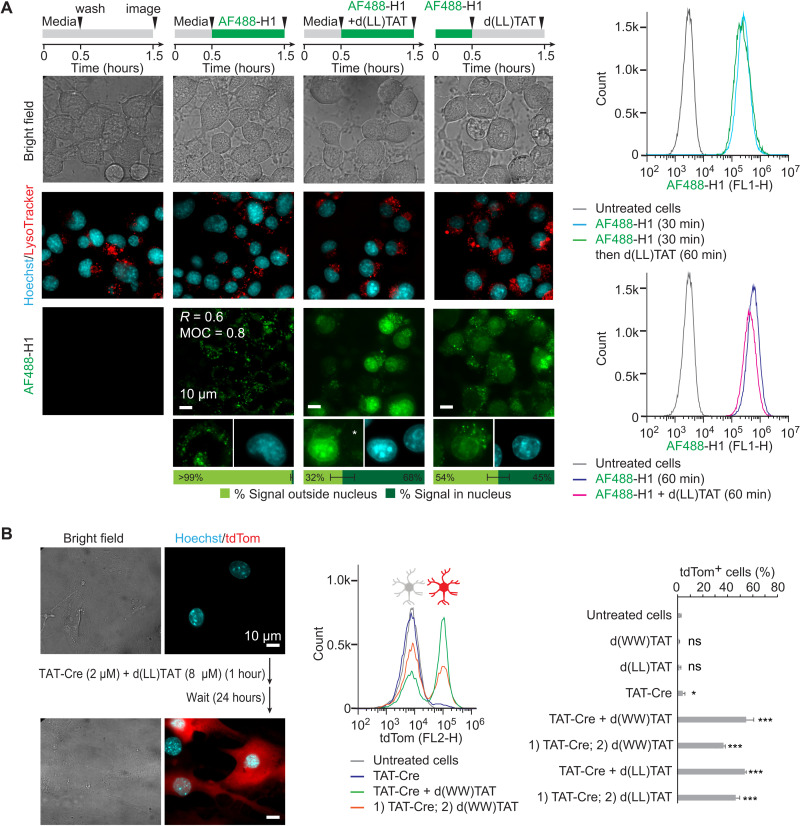
Fig. 2. d(X)TAT constructs successfully deliver the histone H1 and the gene-editing enzyme TAT-Cre recombinase.
(A) Neuro2a cells were exposed to AF488-H1 (0.75 μM) and d(LL)TAT (8 μM) in coincubation or preincubation formats indicated. Representative bright-field and fluorescence images are provided, with AF488-H1 being pseudocolored green and Hoechst being pseudocolored cyan (bleed-through is not present as indicated by the white asterisk). For incubation of AF488-H1 alone, Pearson’s colocalization coefficient (R) and Manders’ overlap coefficient (MOC) were determined for colocalization with LysoTracker red. The average AF488-H1 signal intensity inside and outside nuclei for imaged cells is provided as quantified using total integrated intensity within manually drawn regions of interest in ImageJ. These data were obtained from biological N = 2 duplicates, with 100 cells being imaged per experiment. Flow cytometry was performed to show the relative fluorescence intensity of AF488-H1 in each condition. (B) Delivery of Cre recombinase into primary mouse cortical astrocytes. Cells only express the red fluorescent protein tdTom upon excision of a LoxP-Stop signal by Cre recombinase. Representative bright-field and fluorescence images (tdTom pseudocolored red and Hoechst pseudocolored cyan) and flow cytometry analysis of astrocytes exposed to TAT-Cre, with and without d(X)TAT peptides, are provided. Cells are analyzed 24 hours following Cre delivery to allow for tdTom expression. Delivery was performed using either the co- or preincubation format described in (A). The data represented are the averages and corresponding SDs of biological N = 2 duplicates. *P ≤ 0.05 and ***P ≤ 0.01. ns, not significant.