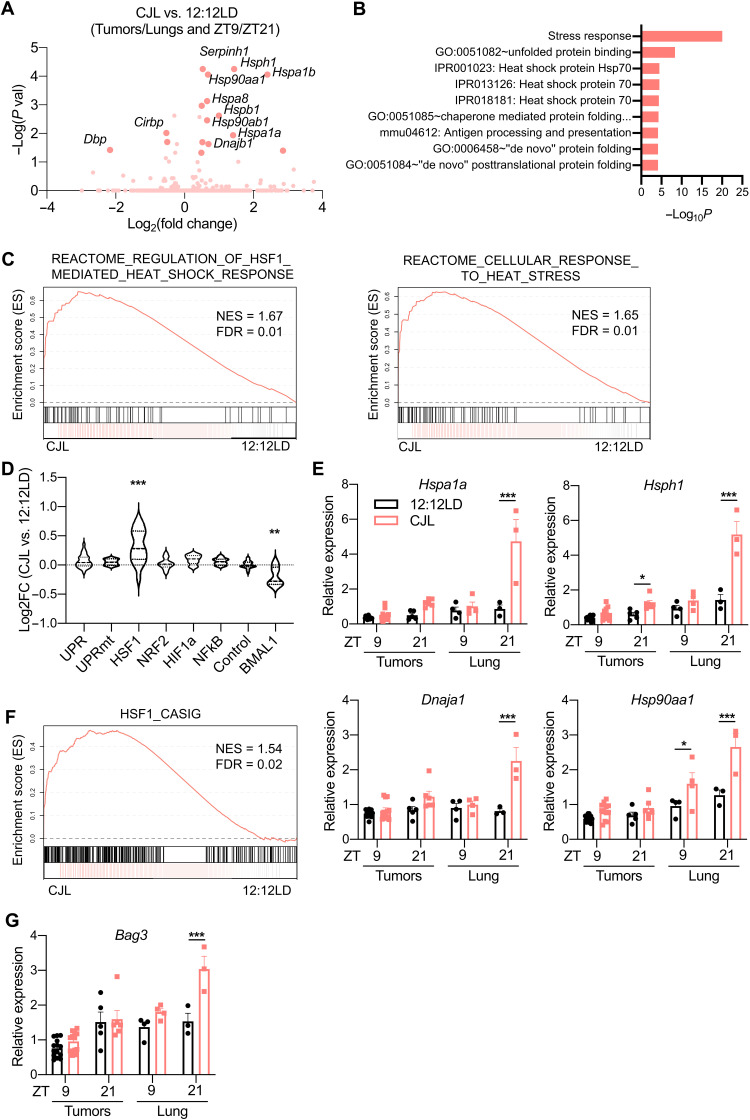
Fig. 4. CJL enhances expression of the HSF1-mediated heat shock response and cancer signature in KrasG12D-driven lung tumor model.
Five weeks after infection with lentivirus-Cre, K mice were placed in either 12:12LD or CJL for 20 weeks. (A) Volcano plots of differentially expressed genes between 12:12LD and CJL by DESeq2 analyses for all samples (tumors + lungs), taking time of collection (ZT9/21) as confounding factor. (B) DAVID analyses on the differentially expressed genes by DESeq2 in all samples between 12:12LD and CJL, taking time of collection as confounding factor. Only terms with false discovery rate (FDR) < 0.25 are shown. (C) GSEA plots for the cellular response to heat stress and HSF1-mediated heat shock response reactome gene sets applied to samples (lungs + tumors) from CJL versus 12:12LD-housed K mice. NES, normalized enrichment score. (D) Activation of stress response pathways by CJL in lungs and tumors, independently of collection time, revealed by grouped fold change for transcripts established as selective targets of each stress pathways (49) or “BMAL1 pathway” (43). **P < 0.01 and ***P < 0.001 by one-way ANOVA with Dunnett’s multiple comparison test. (E and G) Gene expression normalized to U36b4 measured by quantitative real-time PCR; T, tumors; L, lungs. Data represent means ± SEM. n = 14 and 12 for tumors collected at ZT9 in 12:12LD and CJL, respectively; n = 5 and 6 for tumors collected at ZT21 in 12:12LD and CJL, respectively; and n = 3 to 4 for lung samples. *P < 0.05, **P < 0.01, and ***P < 0.001 by two-way ANOVA post hoc Bonferroni test. (F) GSEA plot for the gene set representing the HSF1-CaSig network applied to samples (lungs + tumors) from CJL versus 12:12LD-housed K mice.