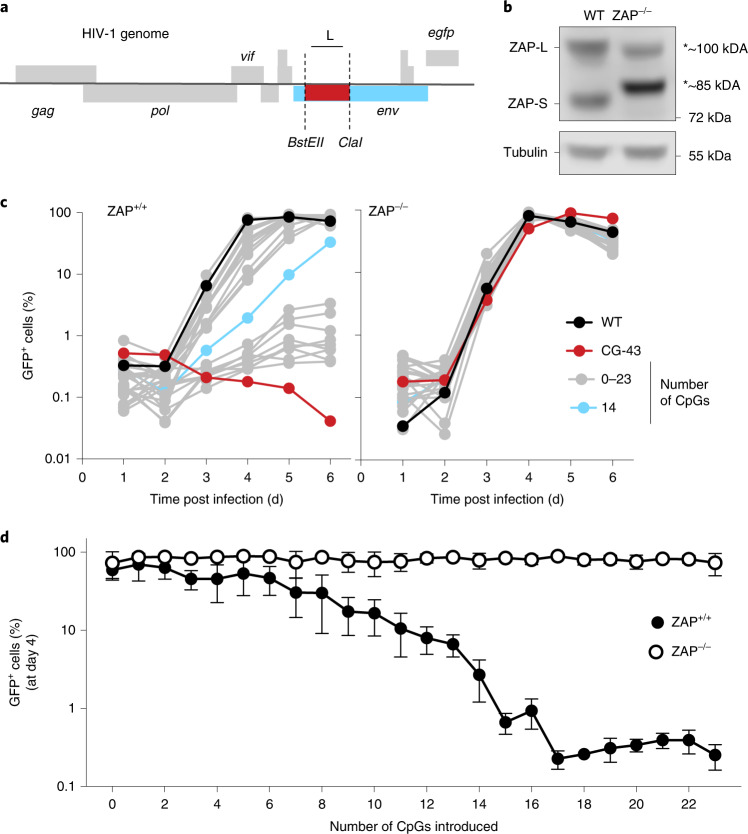
Fig. 1. CpG dinucleotide numbers and HIV-1 replication.
a, Schematic representation of HIV-1 genome containing an EGFP reporter in place of the Nef ORF and introduced restriction sites BstEII and ClaI in the 5′ portion of the env gene. b, ZAP proteins detected by western blotting in unmanipulated and functionally ZAP-deficient human MT4 T cells. The CRISPR lesion results in the expression of truncated non-functional ZAP proteins indicated by asterisks. c, Replication of a collection of HIV-1 mutants containing between 0 and 23 CpG dinucleotides, as well as WT and CG-43 in ZAP-expressing or ZAP-deficient cells. At each day after initial infection, a small sample of cells was collected and the percentage of GFP-positive cells was measured by flow cytometry. d, Percentage of infected cells at day 4 post initial infection across all mutant viruses. Mean ± s.d. from 3 independent experiments; two-way ANOVA for the presence of ZAP (column factor) P < 0.0001, number of CpG (row factor) P < 0.0001. Šídák’s multiple comparisons test was used to calculate adjusted P values between ZAP+/+ and ZAP−/− groups, comparisons of virus mutants with more than 3 CpG display P(adj) < 0.05.