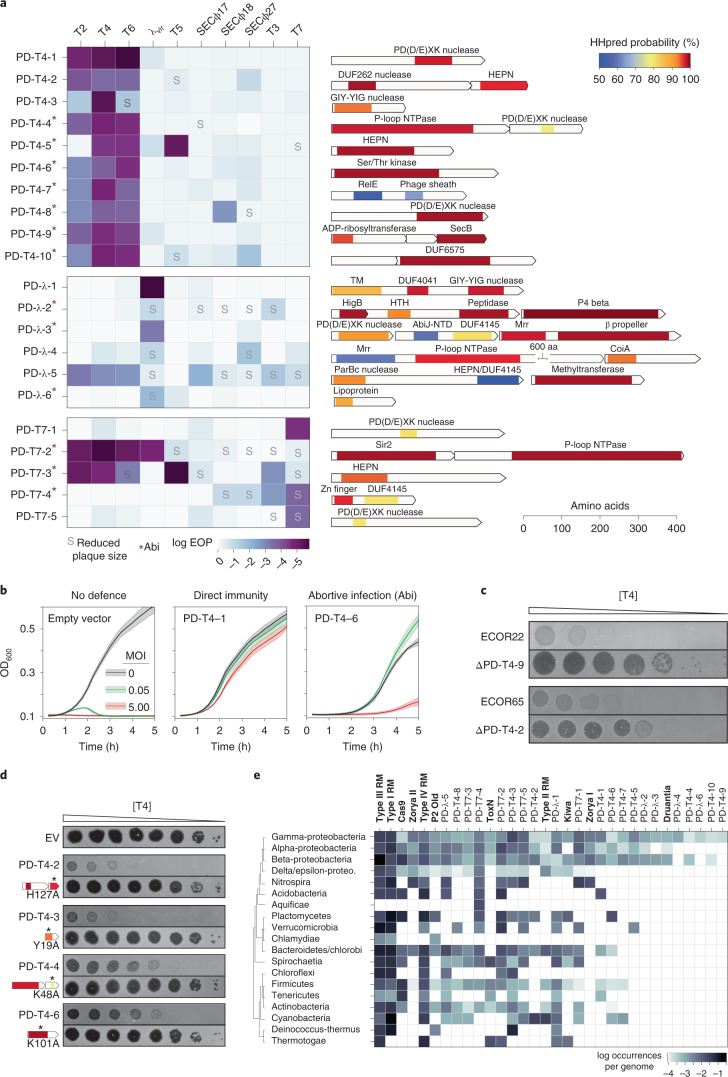
Fig. 3. Summary and annotation of 21 previously uncharacterized defence system loci.
a, Left: each defence system was cloned into a low-copy plasmid with its native promoter and the EOP tested for a panel of ten phages. Darker colours indicate a higher level of protection. Systems leading to smaller plaque sizes are noted with an ‘S’, and systems that protect via an Abi mechanism are indicated with an asterisk. Right: for each defence system identified, the operon structure and predicted domain composition of each component are shown. Shaded regions correspond to domain predictions using HHpred, summarized by association to the PFAM clan, with short descriptions at the top. TM, transmembrane domain; HTH, helix-turn-helix. b, Bacterial growth in the presence of phage at MOIs of 0, 0.05 or 5. Robust growth at MOI 0.05 but not at MOI 5 indicates an Abi mechanism. Lines represent the mean of three technical replicates, with shaded regions indicating s.d. (see Extended Data Fig. 3 for extended MOI data). c, Plaquing of T4 on E. coli isolates ECOR22 and ECOR65 or the isogenic defence system deletions. Dilutions were done on two different plates and images combined for presentation. d, Plaquing of T4 on strains harbouring the indicated defence system or isogenic site-directed mutants of predicted domains. Asterisks indicate approximate location of mutations made. e, Instances of homologues of defence systems by bacterial class, sorted by number of instances, descending from left to right. Known systems are listed in bold for comparison to newly identified systems.