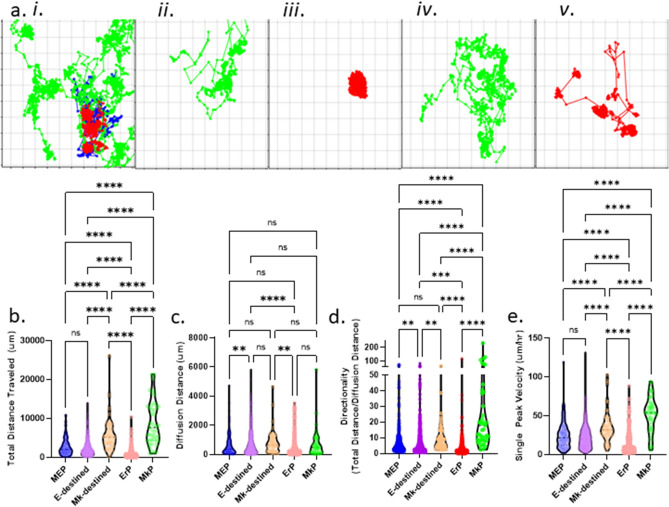
Figure 3.
Motility is a differential behavioral phenotype between progenitor cells undergoing state transitions. (a) Corresponding representative trajectory maps for each colony output. Cell trajectories were generated based on the x,y coordinates of cells in time-lapse CFU assays. Blue trajectories represent MEPs, red trajectories represent E-destined cells, and green trajectories represent Mk-destined cells. Italic roman numerals correspond with Fig. 2a. (b) Total distance traveled by cell state. Total distance was measured as the sum of the distance traveled by an individual cell between time points for the entire lifetime of the bipotent MEP (blue; n = 188), E-destined (purple; n = 964), Mk-destined (orange; n = 49), committed ErP (red; n = 505), and committed MkP (green; n = 47) cell. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns not significant. (c) Diffusion distance traveled by cell state. Diffusion distance was measured as the shortest distance between the starting and ending position of the bipotent MEP (blue; n = 188), E-destined (purple; n = 964), Mk-destined (orange; n = 49), committed ErP (red; n = 505), and committed MkP (green; n = 47) cell at the end of their lifespan. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns not significant. (d) Directionality of motility by cell state. Directionality was calculated by the ratio of total distance over diffusion distance of bipotent MEP (blue; n = 188), E-destined (purple; n = 964), Mk-destined (orange; n = 49), committed ErP (red; n = 505), and committed MkP (green; n = 47). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns not significant. (e) Peak velocity by cell state. Peak velocity is reported as the single highest distance traveled between two time frames over the time between frames for the bipotent MEP (blue; n = 188), E-destined (purple; n = 964), Mk-destined (orange; n = 49), committed ErP (red; n = 505), and committed MkP (green; n = 47) cell. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns not significant.