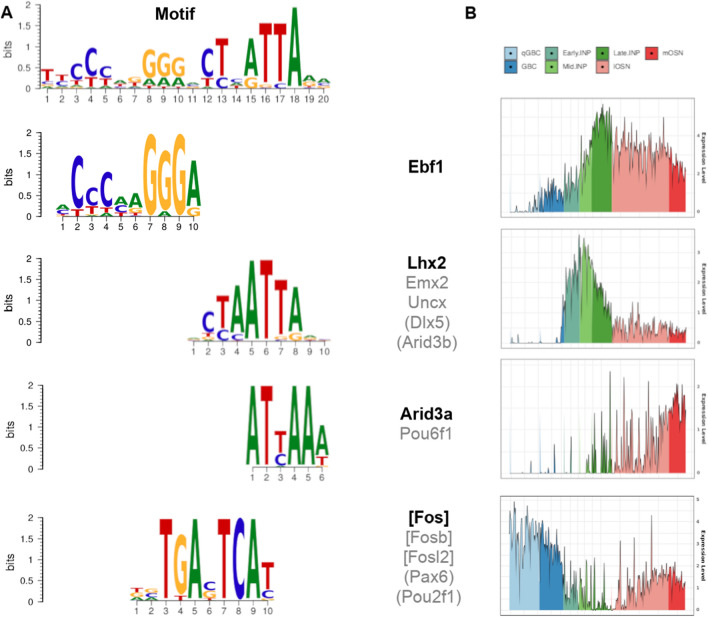
Figure 3.
Greek islands binding motifs and representative pseudotime courses of respective TFs. (A) Binding motifs found in Greek islands. The top row shows the motif found de novo. In the rows below, known binding sites of transcription factors that partly align with the de novo motif in different sites (TOMTOM) and/or are enriched in Greek islands (AME). The transcription factors given in square brackets refer to the TFs found only by Tomtom alignment whereas the round brackets refer to TFs found only by forward motif search (see SFigure 6). (B) Single cell pseudotime courses of the TFs in bold print show characteristic trends along OSN differentiation (y-axis is a natural log of normalized counts). The grouping of the TFs in gray follows their pseudotime profile. It agrees with their grouping according to binding sites, except for Pax6 and Pou2f1 that have a homeobox domain like Lhx2.