Figure 2.
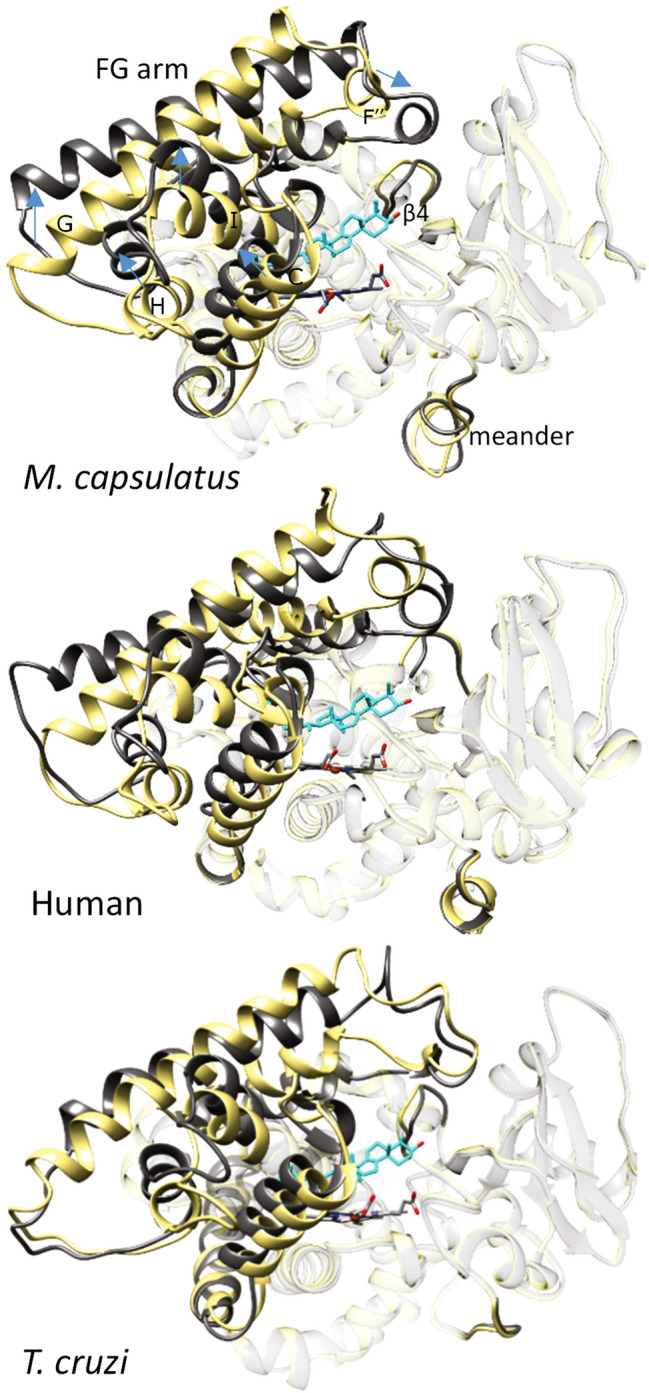
Substrate-induced conformational switch is similar in bacterial and eukaryotic CYP51s. Overlaid structures of substrate-bound (grey) and substrate-free (khaki) CYP51 orthologs. M. capsulatus (7SNM and 6MI0, respectively, rmsd of Cα of 2.44 Å), human (6UEZ and 4UHI, rmsd of Cα of 1.98 Å), and T. cruzi (6FMO, molecules A and D, rmsd of Cα of 1.83 Å). The orientation is about the same (upper P450 view). The directions of the changes are outlined with blue arrows on the M. capsulatus CYP51 structure. The carbon atoms of lanosterol (M. capsulatus and human) and obtusifoliol (I105F T. cruzi) are colored in cyan. The ribbons of the parts of the molecules that are not involved in the conserved conformational change20,21 (7SNM: rmsd of Cα of 0.77) are transparent. For comparison, the ligand-free and detergent-bound M. capsulatus CYP51 structures (6MI0 vs. 6MCW) have RMSD of Cα of 0.37 Å.
