Abstract
Altering plant water use efficiency (WUE) is a promising approach for achieving sustainable crop production in changing climate scenarios. Here, we show that WUE can be tuned by alleles of a single gene discovered in elite maize (Zea mays) breeding material. Genetic dissection of a genomic region affecting WUE led to the identification of the gene ZmAbh4 as causative for the effect. CRISPR/Cas9-mediated ZmAbh4 inactivation increased WUE without growth reductions in well-watered conditions. ZmAbh4 encodes an enzyme that hydroxylates the phytohormone abscisic acid (ABA) and initiates its catabolism. Stomatal conductance is regulated by ABA and emerged as a major link between variation in WUE and discrimination against the heavy carbon isotope (Δ13C) during photosynthesis in the C4 crop maize. Changes in Δ13C persisted in kernel material, which offers an easy-to-screen proxy for WUE. Our results establish a direct physiological and genetic link between WUE and Δ13C through a single gene with potential applications in maize breeding.
Forward and reverse genetics identified the abscisic acid catabolism gene ZmAbh4 as a major link between variation in water use efficiency and carbon isotope discrimination through the control of stomatal conductance.
Introduction
Water use efficiency (WUE) is a central topic in crop research that is focused on securing high yield with reduced water use (Bertolino et al., 2019). The amount of biomass production of a plant per unit transpired water, defined as whole-plant water use efficiency (WUEplant), can be increased by enhancing the efficiency of photosynthetic processes and by limiting transpiration through stomata. WUEplant is highly related to intrinsic WUE (iWUE), defined as the ratio of CO2 assimilation to stomatal conductance, and thus stomatal conductance is an important source of natural genetic variation in WUEplant (Leakey et al., 2019). Most improvements in iWUE and WUEplant have been linked to changes in stomatal characteristics and sensitivity to the phytohormone abscisic acid (ABA; Leakey et al., 2019; Mega et al., 2019; Yang et al., 2019). ABA is known to be a key regulator of stomatal conductance with a major role in restricting transpiration in response to drought, cold, and salt stress (Sah et al., 2016; Venzhik et al., 2016).
We previously identified two maize (Zea mays) lines, recurrent parent RP and near-isogenic line NIL B, that differ in WUEplant, iWUE, stomatal conductance, stomatal density, leaf ABA concentrations, growth sensitivity to drought, and kernel carbon isotope discrimination (Δ13C; Gresset et al., 2014; Avramova et al., 2019). All of these traits were associated with a 55 Mb introgression on chromosome 7 differing between RP and NIL B. RP, the recurrent parent of the introgression library, is an elite Southeastern-European dent (Z. mays indentata) line with high drought tolerance. The introgression of NIL B originates from a drought-sensitive flint (Z. mays indurata) donor parent of the introgression library used for kernel Δ13C mapping (Gresset et al., 2014). The close genetic similarity of NIL B and RP (Avramova et al., 2019) made the two genotypes an excellent starting point to unravel the genetic basis and causal connection between the traits. While the influence of ABA on stomatal conductance and the connection to WUEplant and drought sensitivity has been shown in multiple species (Zhang, 2014; Leakey et al., 2019), the relationship between Δ13C and WUEplant in C4 plants requires further research (Eggels et al., 2021).
The Δ13C of dry matter is influenced by the extent of discrimination against the heavier isotope 13C relative to 12C during carbon fixation processes. Δ13C depends strongly, but not exclusively, on the ratio of intercellular (Ci) to ambient (Ca) CO2 concentration (Farquhar, 1983), which can be derived directly from iWUE. Due to the close relationship between Δ13C and iWUE and because it is a relatively easy parameter to measure in a large number of genotypes, Δ13C of plant biomass has been established as a proxy for iWUE and WUEplant in C3 plants (Condon et al., 2004; Chen et al., 2011). In C4 plants, however, the initial carbon fixation by phosphoenolpyruvate carboxylase (PEPC) and the compartmentalization of photosynthesis into two cell types (which is associated with the leakage of CO2 from bundle sheath cells back to mesophyll cells [leakiness]) commonly reverse and weaken the correlation of Δ13C and iWUE compared to C3 plants (von Caemmerer et al., 2014).
In the present study, we show that in the C4 crop maize, the single gene ZmAbh4, encoding an ABA 8′-hydroxylase, can alter both WUE and Δ13C. Through genetic dissection of a 55 Mb region on chromosome 7 associated with both traits, we identified ZmAbh4 as the causative gene and verified its effect using targeted knockouts. Furthermore, we demonstrate that the association of WUEplant and Δ13C in the studied maize lines originates from the correlation of photosynthetic Δ13C with the ratio of intercellular and ambient CO2 levels (Ci/Ca). Our findings point to the potential of using Δ13C as a proxy for WUE in maize, and identify a gene with potential applications for maize breeding.
Results
Both WUE and Δ13C are modulated by ZmAbh4
The two maize lines, the recurrent parent RP and NIL B, share 97% genome identity and differ in a 55 Mb introgression on chromosome 7 associated with changes in WUEplant, iWUE, and differences in kernel carbon isotope discrimination (Δ13CKernel) of field-grown plants (Avramova et al., 2019; see also Supplemental Figure S1, A–C). The lower iWUE in NIL B is explained by its higher stomatal conductance (gs), whereas it only has a minor increase in the assimilation rate compared to RP (Supplemental Figure S1, D and E). Additionally, leaf ABA levels are lower in NIL B (Supplemental Figure S1F) and are a likely cause for the higher stomatal conductance of this line, as ABA negatively affects stomatal aperture (Mittelheuser and Van Steveninck, 1969). We used both maize lines as a starting point to identify genes responsible for the observed differences in WUE and Δ13C.
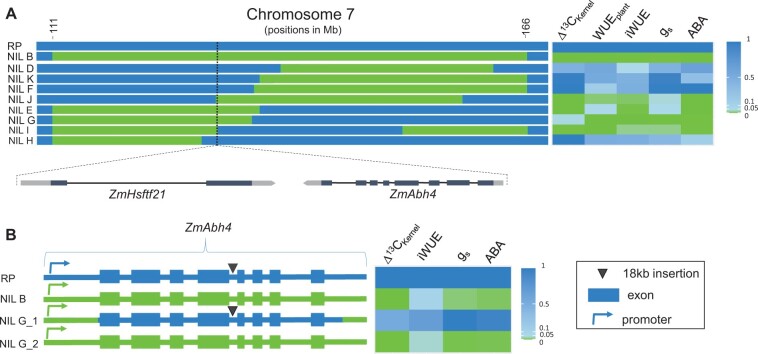
In a mapping population consisting of 1,306 plants derived from the backcross of NIL B to RP, 342 recombinants were identified and eight homozygous lines (named NIL D-K; Figure 1A) comprising recombinant breakpoints within the target region were developed (Supplemental Table S1). With respect to the traits gs, iWUE, WUEplant and Δ13CKernel, the eight near-isogenic lines (NILs) either resembled RP or NIL B (Figure 1A;Supplemental Table S2A). The four NILs E, G, J and I share a ∼200 kb segment (129.798–129.995 Mb) associated with all the measured traits. This chromosomal segment spans two annotated genes in the maize B73 reference genome (Zm-B73-REFERENCE-GRAMENE-4.0; Jiao et al., 2017): the ZmHsftf21 gene (Heat shock factor transcription factor 21, also known as Heat shock factor protein 4) and the ZmAbh4 gene (ABA 8′-hydroxylase 4, also known as ZmABA8Ox3a). ZmHsftf genes are known to play a role in abiotic stress responses (Zhang et al., 2020), and ZmAbh genes take part in ABA catabolism (Okamoto et al., 2009). A functional difference between the ZmAbh4 allele of RP and NIL B might explain the observed variation in leaf ABA levels.
Figure 1.
Genetic dissection of a 55 Mb region on chromosome 7 identifies ZmAbh4 as modulating WUE and Δ13C in maize. A, Genomic composition of nine near-isogenic maize lines (NILs) and the recurrent parent (RP) in the region between 111 and 166 Mb on chromosome 7. Illustrated are homozygous introgressions from a donor parent in the genetic background of the recurrent parent. Dotted line marks the 200 kb region (129.798–129.995 Mb), where the genes ZmHsftf21 and ZmAbh4 are located. Breakpoint positions for all NILs are given in Supplemental Table S1. B, ZmAbh4 gene structure in RP and three NILs. The RP allele of the gene (blue) contains an 18 kb insertion in intron 4 (marked by a triangle), which is not present in the NIL B allele (green). The respective properties of the NILs compared to RP (two-sided Dunnett’s test) in terms of kernel carbon isotope discrimination (Δ13CKernel), whole plant WUE (WUEplant), intrinsic water use efficiency (iWUE), stomatal conductance (gs), and ABA leaf concentrations were assessed and P-values are presented as heatmaps (P < 0.05 in green). Means ± SD and n are presented in Supplemental Table S2.
To further dissect the introgressed chromosome region, a backcross of NIL G to RP was performed and resulted in the identification of NIL G_1 and NIL G_2, which differ only in the ZmAbh4 allele (Figure 1B), while both carry the NIL B allele of ZmHsftf21 (Supplemental Figure S2). The ZmAbh4 allele in NIL G_1 originates from RP from approximately 50 bp before the translation start site until the end of the 3′ UTR (Figure 1B). The promoter and any other proximal upstream regulatory elements, which could potentially influence gene expression, originate from NIL B in both lines (Supplemental Figure S2). Phenotyping of NIL G_1 and NIL G_2 demonstrated that the higher ABA, lower gs, higher iWUE, and Δ13CKernel are caused by the RP ZmAbh4 allele (Figure 1B; Supplemental Table S2B). Genomic sequencing of chromosomal DNA fragments covering the ZmAbh4 locus (Supplemental Data Set S1) identified an 18 kb insertion between the fourth and fifth exon of ZmAbh4 that is not present in the NIL B allele (Figure 1B) and might impair the formation of the mature transcript. Additional polymorphisms within the gene and proximal sequences mainly lead to conservative amino acid exchanges and do not affect domains known to be important for the function of the gene (Supplemental Table S3).
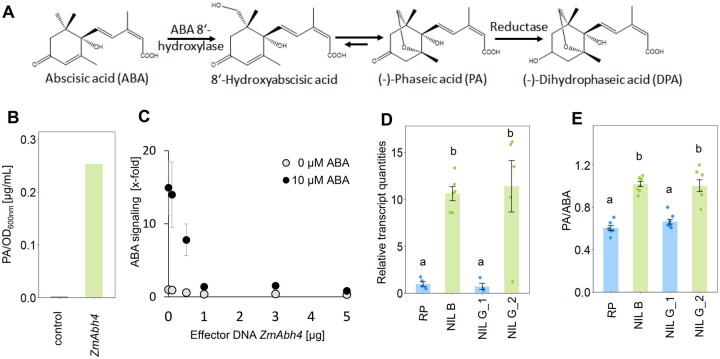
ZmAbh4 was annotated as encoding an ABA 8′-hydroxylase (ABH; Jiao et al., 2017). ABHs are key enzymes of the predominant catabolic pathway of ABA that catalyze the first step of the reaction, the 8′-hydroxylation of ABA to 8′-hydroxyabscisic acid, which in turn isomerizes to phaseic acid (PA) and is further catabolized to dihydrophaseic acid (DPA; Figure 2A; Krochko et al., 1998).
We demonstrated the functionality of the ZmAbh4-encoded enzyme as an ABA 8′-hydroxylase by detecting elevated PA levels in the supernatant of ZmAbh4-expressing yeast incubated in the presence of ABA (Figure 2B). Consistent with an ABA catabolic function of ZmABH4, ABA-induced signaling in Arabidopsis thaliana protoplasts was efficiently decreased (90%) by expressing the hydroxylase (Figure 2C). The cDNAs of the ZmAbh4 alleles of NIL B and RP were able to decrease ABA-induced signaling to a similar extent, which suggests that polymorphisms found in the exons did not contribute to the observed phenotype.
Figure 2.
ZmAbh4 encodes an ABA 8′-hydroxylase that modulates ABA catabolism in the maize leaf. A, Main catabolic pathway of ABA. B, ABA 8′-hydroxylase encoded by the NIL B ZmAbh4 allele converts ABA to PA, as shown by ZmAbh4 expression in yeast. Concentration of PA in the supernatant of the yeast culture was normalized to the OD600nm of the culture at harvest. In the control (yeast without ZmAbh4 expression), PA was below the detection limit. C, ABA signaling in Arabidopsis protoplasts that were transiently transfected with varying amounts of ZmAbh4 (NIL B allele) effector DNA with equal amounts of total DNA (5 µg, adjusted with vector control) and incubated in the presence or absence of 10 µM ABA. ABA signaling is given as ABA-dependent luciferase induction relative to the value without exogenous ABA and effector DNA. Means ± SE (n = 6 transfections of 105 protoplasts each). Transfection with 3 µg cDNA of the RP and NIL B alleles of ZmAbh4 resulted in equal reductions in ABA signaling for both alleles. D, Transcript levels of ZmAbh4 in the leaves of plants at the V5 stage, measured with primers spanning intron 4. Means ± SE (n = 4–6 plants). E, Ratio of leaf 5 PA and ABA concentrations in the last developed leaves at the V5 stage. Means ± SE (n = 7–8 plants). Significant differences (LSD test) in (D) and (E) are marked with different letters.
The ZmAbh4-mediated decrease in ABA signaling is dependent on ABA-hydroxylase levels (Figure 2C), which might be compromised in RP and NIL G_1 because of the insertion in the ZmAbh4 allele. The insertion is located between exons 4 and 5 of the primary transcript and likely impairs its maturation. Hence, we analyzed the correct splicing of the ZmAbh4 transcript in leaves of RP and NIL B using primer pairs spanning each of the seven introns (Supplemental Table S4). At exon 5, the mature transcript levels of RP were close to the detection limit, and low transcript levels were also maintained at the subsequent exons compared to NIL B (Supplemental Figure S3). Transcript levels in NIL G_1 and NIL G_2 showed similar patterns to RP and NIL B, respectively, when measured with the same set of primers spanning the fourth intron (Figure 2D). Examining the 18 kb insertion in this intron using an established annotation pipeline (Ou et al., 2019) led to the identification of a non-autonomous Helitron transposon (sequence ontology SO:0000544) +39 and +41 bp upstream of the donor splice site and coding exon end, respectively. Transposable elements have been found to commonly disrupt transcription (Han et al., 2004). The findings pinpoint the near loss of ZmAbh4 function possibly resulting from this Helitron insertion as the cause for higher WUEplant and Δ13C in RP.
The impaired expression of ZmABH4 in RP was expected to decrease ABA catabolism. Indeed, elevated ABA levels and decreased PA/ABA ratios were detected in the leaves of all genotypes carrying the RP allele of ZmAbh4 compared to genotypes carrying the NIL B allele (Figure 2E;Supplemental Table S2). Elevated leaf ABA levels can explain the lower gs leading to higher iWUE, WUEplant and higher Δ13C in the genotypes carrying the RP allele compared to those carrying the NIL B allele of the gene (Figure 1; Supplemental Table S2). We, therefore, conclude that ZmAbh4 controls ABA levels in the leaves and causes the observed differences in gs, iWUE, WUEplant and Δ13CKernel in the analyzed genotypes (Figure 1; Supplemental Figure S1).
Targeted mutations in the coding region of ZmAbh4 cause changes in WUE and Δ13C
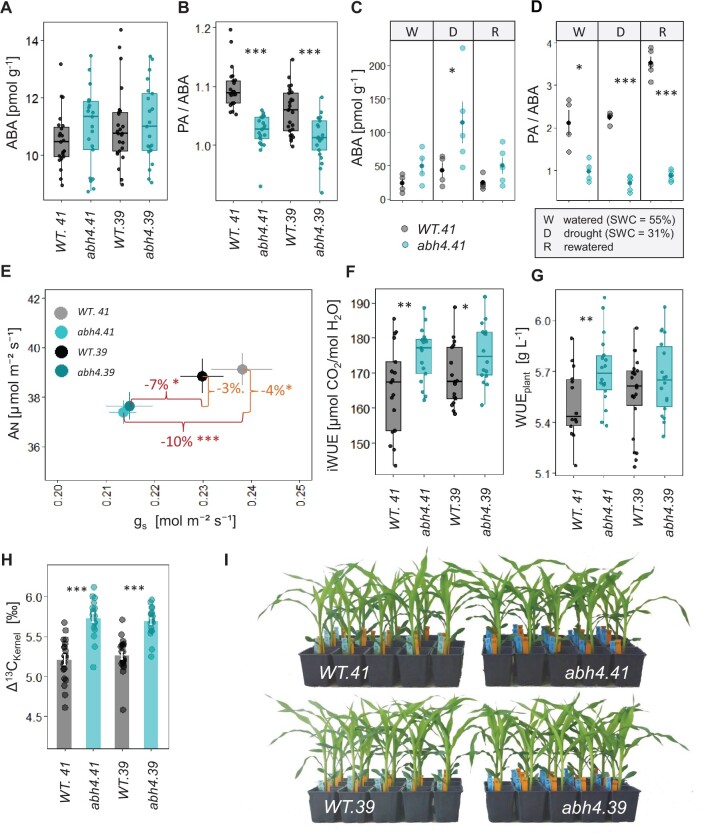
To investigate whether targeted mutations into the coding sequence of ZmAbh4 cause a similar phenotype to that of the RP allele, we generated CRISPR/Cas9 mutants in a different genomic background (inbred line B104). Two independent CRISPR/Cas9 mutants, abh4.41 and abh4.39, showed a 157 bp deletion and a 1 bp insertion in ZmAbh4, respectively, both resulting in frameshifts and premature stop codons (Supplemental Figure S4, A and B). The non-functionality of the mutant alleles was verified using the established yeast assay (Eggels et al., 2018; Supplemental Figure S4C).
The knockout of ZmAbh4 in the mutants was expected to affect the concentration of ABA and its catabolites in maize leaves. The ABA content in the leaves of well-watered plants was similar in the mutants and respective wild types (WTs; Figure 3A). However, the ratio of the catabolite PA to ABA was clearly reduced in both abh4 mutants (Figure 3B), indicating reduced ABA 8′-hydroxylation. When subjected to mild drought stress followed by rewatering, significant differences between abh4.41 and WT.41 were found in the PA/ABA ratio under both conditions and also for ABA levels under drought (Figure 3, C and D).
Figure 3.
Functional inactivation of ZmABH4 by CRISPR/Cas9 alters the ratio of PA to ABA and water relations in maize. A–D, Content of ABA, and the ratio of the first stable product of ABA 8′-hydroxylation, PA, relative to ABA in fully developed leaves measured (A and B) in leaves of well-watered plants (n = 21–25 plants), (C and D) in leaves of plants grown under either well-watered conditions, mild drought stress, or mild stress with subsequent rewatering (n = 4–5 plants, shown are means ± SE and individual data points). E, Assimilation rate (AN) over stomatal conductance (gs) of well-watered plants (n = 16–17 plants, mean ± SE). The percentage decrease in the mean value of the mutant relative to its WT is shown for AN and gs. F, Intrinsic water use efficiency (iWUE) calculated from data shown in E (n = 16–17 plants). G, WUEplant of plants grown under progressive drought to determine the dry weights plants are capable of attaining with a given amount of water, n = 14–22 plants. H, Carbon isotope discrimination of kernels (Δ13CKernel), n = 18–22 plants. Bars show means ± SE. Boxplots with center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range. One-sided Student’s t test between the abh4 mutant and respective WT with ***P < 0.001, **P < 0.01, *P < 0.05. I, Phenotypes of well-watered mutant and WT plants.
Consistent with observations in the NILs, the two abh4 mutants showed a significant reduction in gs, which was associated with only a minor reduction in the apparent rate of CO2 assimilation (Figure 3E, P < 0.05 for the comparison abh4.41 to WT.41, P < 0.1 for abh4.39 to WT.39). Therefore, iWUE was increased in the abh4 mutants relative to their respective WTs (Figure 3F). This increase in iWUE translated to increased WUEplant for abh4.41, and a trend towards increased WUEplant was also observed for abh4.39, although it was below the significance threshold (0.05 < P < 0.1; Figure 3G). We, therefore, conclude that a significant increase in WUE (iWUE and WUEplant) of maize can be achieved by the loss-of-function of a single gene, despite the polygenic nature of the trait.
The higher Δ13C detected in kernels of the abh4 mutants compared to their WTs (Figure 3H) is further evidence for the connection of iWUE and WUEplant with Δ13C in maize. Overall, the effects on gs, WUE, and Δ13C of targeted mutations in the coding sequence of ZmAbh4 led to a similar phenotype to the impaired formation of the complete transcript in the RP allele and were stable across the two different genomic backgrounds examined in this work.
No consistent differences in growth were observed for RP and NIL G_1, carrying the near loss-of-function ZmAbh4 allele, compared to NIL B or NIL G_2, respectively, nor for the abh4 mutants compared to their WTs (Supplemental Figure S5, A and B; Figure 3I. Additionally, no consistent disadvantage regarding germination rate and yield components such as kernel number per cob and thousand kernel weight could be observed due to the disruption of ZmAbh4 function (Supplemental Figure S5, C–J).
Δ13C of leaves and kernels reflects differences in photosynthetic carbon isotope discrimination caused by stomatal conductance
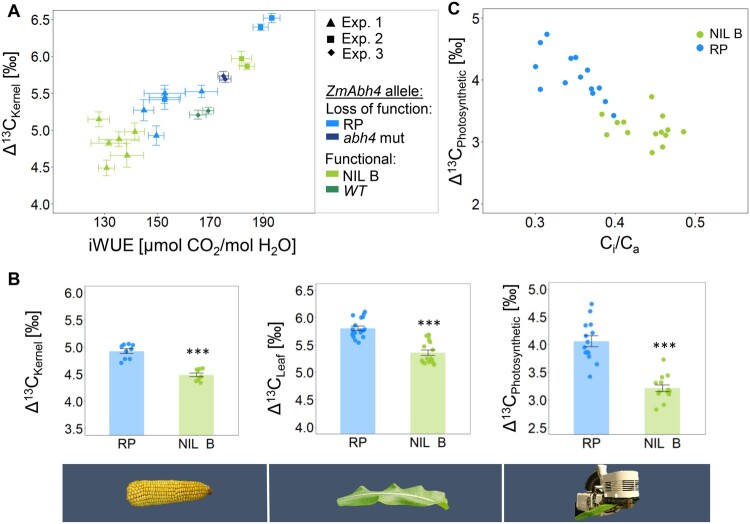
Analyses of NILs and mutants demonstrated that the higher iWUE caused by the loss of functional ZmAbh4 transcript is accompanied by higher Δ13CKernel. Over the different genotypes and experiments, a positive relationship between iWUE and Δ13CKernel was observed under the assumption that the carbon isotopic composition of air was comparable in all experiments (Figure 4A). Online measurements of the photosynthetic Δ13C of RP and NIL B showed that differences in photosynthetic Δ13C underlie the link between the observed differences in gas exchange and Δ13CKernel. Photosynthetic Δ13C was significantly lower in NIL B compared to RP, which is consistent with the lower Δ13C measured in dried leaf material at different developmental stages and in kernel material from NIL B (Figure 4B; Supplemental Figure S6B). This strongly suggests that these differences in Δ13C during photosynthesis persist throughout the plant lifecycle and are causative for the differences in Δ13C in leaf and kernel material.
Figure 4.
Carbon isotope discrimination (Δ13C) is physiologically linked to WUE. A, Association of kernel Δ13C (Δ13CKernel) and intrinsic WUE (iWUE) measured at stage V5/6 in different experiments (Exp. 1–3) and genotypes (data from Figures 1, 3, and 4B. Data from near-isogenic lines (NILs) carrying the recurrent parent (RP) ZmAbh4 allele or the NIL B allele, and from abh4 CRISPR mutants (abh4 mut) and their wild types (WT) are shown). Plants were grown in a greenhouse for Exp. 1 and 3 and in a growth chamber for Exp. 2. Shown are means ± SE for each genotype per experiment. B, Δ13C derived from isotopic compositions of dry matter (Δ13CKernel, n = 9 plants; Δ13CLeaf, n = 17–18 plants) and photosynthetic Δ13C (Δ13CPhotosynthetic, n = 14 plants) from online gas exchange measurements for RP and NIL B. Bars indicate mean ± SE. Significant differences compared to RP based on two-sided Student’s t test are indicated by ***P < 0.001. C, Relationship between Δ13CPhotosynthetic and concurrently measured ratio of intercellular to ambient CO2 concentration (Ci/Ca) in RP and NIL B.
The 20% lower photosynthetic Δ13C in NIL B compared to RP was associated with a simultaneously 25% higher ratio of intercellular-to-ambient CO2 concentration (Ci/Ca) observed in NIL B, demonstrating a negative dependence of Δ13C on Ci/Ca (Figure 4C). Ci/Ca and iWUE are inversely linked because they are contrastingly influenced by stomatal conductance, so the observed relationship of both photosynthetic and kernel Δ13C with iWUE is positive (Supplemental Figure S6A; Figure 4A). A negative relationship of Δ13C and Ci/Ca, i.e. positive relationship of Δ13C and iWUE, is expected if the extent of leakage of CO2 from bundle sheath to mesophyll cells (leakiness) is less than 0.37 because of the enrichment of 13C associated with the initial carbon fixation step (Farquhar, 1983). Mean estimates of leakiness based on the simplified Δ13C model (Farquhar, 1983) were below this threshold and were lower in NIL B (0.27 ± 0.02) than in RP (0.34 ± 0.04). In addition to the effect of Ci/Ca, such differences in leakiness might thus have contributed to the differences in Δ13C between the two genotypes.
In summary, iWUE and thus Ci/Ca drive changes in Δ13C during photosynthesis in RP and NIL B, which are reflected in Δ13C values of leaves and kernels. The decrease in iWUE in NIL B is driven by the higher stomatal conductance, which does not cause a corresponding increase in assimilation, because the assimilation rates of the NILs are close to saturation at ambient CO2 concentrations of 400 ppm (Supplemental Figure S7A). Measuring chlorophyll content and the activities of the carboxylating enzymes PEPC and Rubisco did not give any indications of the involvement of additional factors linked to assimilation (Supplemental Figure S7, B–E). Therefore, we conclude that stomatal conductance is the main factor explaining the simultaneous changes in iWUE and Δ13C in maize.
Discussion
We demonstrated that, although WUE is a complex quantitatively inherited trait (Wang et al., 2018), the gene ZmAbh4 can induce significant changes in stomatal conductance and WUE. ZmAbh4 alleles show environmental plasticity and allow for the fine-tuning of WUE depending on the environmental scenario. Both higher and lower WUE can be beneficial for the final crop yield under drought, depending on the severity and duration of the stress (Tardieu et al., 2018). Selecting for either the functional or non-functional allele thus allows breeders to modulate these traits depending on environmental conditions. The co-occurrence of a Helitron transposon and of altered transcript levels for the RP allele measured using primers downstream of this transposon (Figure 2D, Supplemental Figure S3) provides strong support for the notion that the 18 kb insertion in the fourth intron is likely the causative mutation for the phenotypic differences between the RP and NIL B genotype. This insertion is molecularly detectable as a natural variant of the gene. Screening of the publicly available genome assemblies of 25 American NAM (Nested Association Mapping) parents (Hufford et al., 2021) by BLAST analysis and a set of 30 American and European lines by PCR (Supplemental Figure S8) identified only two genotypes (Mo17 and EC49A) carrying this allele. As both alleles of ZmAbh4 were identified in elite genetic material (Gresset et al., 2014), we hypothesize that the advantageous effect of the mutation causing increased WUE via reduced stomatal conductance is beneficial only in some target environments.
The loss of ZmAbh4 function through CRISPR/Cas9 knockout led to significant differences in ABA content under mild drought stress (Figure 3C) but did not have a significant influence on ABA levels under well-watered conditions (Figure 3A). Similarly, ABA measurements obtained from knockouts of the homologous gene ZmAbh2 showed significant changes between mutant and WT only for drought-stressed plants (Liu et al., 2020). Still, the alteration of PA/ABA ratio under well-watered conditions in the abh4 mutants compared to the respective WT shows that the ZmAbh4 knockouts cause shifts in phytohormone homeostasis even under these conditions (Figure 3B). Further research is needed to determine if the effect of the ZmAbh4 knockout is restricted to specific tissues or cell types within the leaves, which might lead to non-detectable differences at the whole leaf level but still affect the plant’s gs and iWUE, as observed here (Figure 3, E and F).
In addition to modulating ABA homeostasis by knocking out an ABA catabolic gene, altering ABA sensitivity by overexpressing ABA receptors led to an increase in WUE in Arabidopsis (Yang et al., 2016) and wheat (Triticumaestivum; Mega et al., 2019). The knockout of ZmAbh2, a maize homologue of ZmAbh4 also expressed in leaves, enhanced drought tolerance, demonstrating another beneficial effect of modulating ABA catabolism (Liu et al., 2020). Overexpression of the E3 ubiquitin ligase gene ZmXerico1 achieved increased WUE and drought tolerance in maize by influencing ABA homeostasis (Brugière et al., 2017). ZmXerico1 was suggested to regulate the protein stability of ZmABH1 and ZmABH4. In addition to enhanced WUE, its overexpression led to ABA hypersensitivity, as demonstrated by increased suppression of root elongation under ABA treatment in maize and decreased germination rates in Arabidopsis. The five members of the ZmAbh gene family in maize have tissue-specific expression (Vallabhaneni and Wurtzel, 2010), with the highest expression of ZmAbh1 in roots and the highest expression of ZmAbh4 in leaves. We showed that targeting ZmAbh4 directly allows for the fine-tuning of ABA concentrations in leaves without observable undesirable effects on other traits such as germination or growth (Supplemental Figure S5). We conclude that the ZmAbh gene family has potential for crop improvement and is worthy of further exploration.
In crop breeding, one of the major limiting factors for the selection of genotypes with differences in WUE is the lack of high-throughput screening, as measuring both iWUE and WUEplant is time-consuming and labor-intensive. In C3 crops, Δ13C has been established as an efficient proxy for estimating WUE at high throughput and has been implemented in a breeding program (Condon et al., 2004). The association of Δ13C and WUE was suggested to hold true for C4 plants (Henderson et al., 1998; Twohey et al., 2019; Ellsworth et al., 2020), which represent a large proportion of crops in cultivated areas, but direct physiological and genetic evidence for this has been lacking. Here we showed that in maize, the two traits Δ13C and WUE are influenced by a common gene, ZmAbh4. Stomatal conductance was the central trait causing co-variation of iWUE and photosynthetic Δ13C measured during gas exchange, resulting in a positive correlation of WUE and Δ13C derived from dried plant material (leaves, kernels). Differences in leaf and kernel Δ13C reflected differences in the Δ13C during photosynthesis. Our study demonstrates the potential of using Δ13C as a proxy for WUE in maize, yet further research is needed to determine if this relationship between Δ13C and WUE holds across diverse genetic material.
Materials and methods
Plant material
Two previously described (Avramova et al., 2019) maize (Zea mays) lines, RP and NIL B, (genomic background of a dent inbred line belonging to the non-Stiff Stalk heterotic group) and lines originating from a cross between them were used for the experiments. Additionally, CRISPR/Cas9 mutants in the B104 genomic background were studied. B104 is a temperate American inbred line of the Stiff Stalk heterotic group commonly used to obtain transgenic plants (Feys et al., 2018).
Generating recombinant NILs
Crossing NIL B and RP resulted in an F2 mapping population including NILs with recombination events in the introgression on chromosome 7 carried by NIL B (55 Mb). Homozygous lines with different recombinant breakpoints within this target region (Supplemental Table S1) were obtained by self-pollination of selected F2 plants and used for phenotyping. The genomic background of these lines and the lengths of their introgressions were tracked by genotyping with the 600 k Axiom™ Maize Genotyping Array (Unterseer et al., 2014). One of the homozygous lines (NIL G) was further backcrossed to RP, and a plant heterozygous for recombination events at the ZmAbh4 locus was selected in the F2 generation and self-pollinated. After genotyping the progeny of this plant, two homozygous lines were selected: NIL G_1 and NIL G_2. NIL G_2 has the same genotype as the parent NIL G, whereas NIL G_1 differs from NIL G and NIL G_2 only at the ZmAbh4 locus, where it carries the RP instead of the NIL B allele.
Obtaining CRISPR/Cas9 mutants
The genomic sequences of ZmAbh genes of maize inbred line B104 were obtained from MaizeGDB (www.maizegdb.org; Portwood et al., 2019). Two target sites (5'-N20NGG-3') per gene were selected based on the location within the first exons, high-efficiency scores, and low number of unrelated off-targets (Concordet and Haeussler, 2018).
Cloning was performed via Gateway reactions as described (Xing et al., 2014). The plasmids pBUN411, pCBC-MT1T2, pCBC-MT2T3, and pCBC-MT3T4 were gifts from Qi-Jun Chen, China Agricultural University, Beijing, China (Addgene plasmid # 50581 # 50593, # 50594 # 50595; Xing et al., 2014).
CRISPR/Cas9 mutants of ZmAbh4 were generated in the maize inbred line B104 background using four gRNAs targeting ZmAbh4 and ZmAbh1 (CGTGGAACGTGCTTCTGACG and GCCTTGTAATGGAGCGTCCC targeting ZmAbh4, AGGATGTGCGTCTTGAAGAT and GCTGCGCTCGCTCGAGTCCT GGG targeting ZmAbh1). Agrobacterium-mediated transformation was performed at the VIB Center for Plant Systems Biology, Ghent, Belgium. In the T1 generation, two Cas9 null segregants (41 and 39) were selected for detailed analysis. The lines, which originated from different events, carried individual mutations in ZmAbh4 and the WT genotype for all homologous genes. The segregating T2 generation was genotyped and classified into homozygous mutants (abh4.41 or abh4.39) and respective wild types (WT.41 or WT.39) originating from the same cob. Phenotyping was performed in theT2 and T3 generations.
Growth conditions
If not stated otherwise, all phenotyping was conducted on plants grown in the greenhouse (Plant Technology Center, Technical University of Munich, Freising, Germany). Plants that were grown for 2–3 weeks in a growth chamber under previously described conditions (Avramova et al., 2019) were transferred to 8 L of soil in the greenhouse. Environmental conditions in the greenhouse were maintained at 25°C–35°C/18°C–20°C day/night, 40% relative humidity (RH), and light intensity of 600 µmol m−2s−1 supplemented by ceramic metal-halide lamps when needed.
For mild drought stress applied to the CRISPR mutants, 3-week-old plants were transferred to pots with 8 L of sifted soil (B 480, Stender, Germany) in the greenhouse. All plants were well-watered for four additional days before water was withheld for the drought (D) and rewatering (R) groups while the well-watered (W) group received daily watering to 58.5% volumetric soil water content (SWC, volume of water divided by initial soil volume: 4.68 L/8 L). Plants under R conditions were rewatered on the 10th day, and leaves were harvested 5 h later from plants under all conditions. At harvest, group D still experienced mild stress (SWC 31%), while plants in groups W and R did not show stress symptoms (SWC 55%).
Concurrent measurements of photosynthetic Δ13C and gas exchange, recording of A/Ci curves (see below), as well as comparative phenotyping of NIL G_1, NIL G_2, RP and NIL B were performed with plants grown in a growth chamber at the Plant Technology Center (Technical University of Munich, Freising, Germany) with 800 μmol m−2 s−1 light intensity at plant height provided by cool-white fluorescent tubes and warm-white light-emitting diode (LED) bulbs and 60% RH until reaching plant stage V5/6.
For all experiments, plants were arranged in a randomized complete block design. The developmental stages of maize were determined according to https://www.pioneer.com/us/agronomy/staging_corn_growth.html.
To measure Δ13CKernel in field-grown plants, plants were grown in Freising, Germany (48°24′12.2″N, 11°43′22.3″E) in 2019. Supplemental watering was applied when necessary to avoid drought stress. The genotypes under study were part of a trial containing 91 entries that were planted in a randomized complete block design with three replications per entry (genotypes described in this study were present as duplicate entries) and analyzed as described previously (Avramova et al., 2019).
Determination of whole-plant water use efficiency
Assessment of WUEplant was performed as previously described (Yang et al., 2016; Avramova et al., 2019). Plants were grown in the growth chamber for 2 weeks until vegetative stage V3-4, transferred to 10 L watertight pots filled with 8 L of sifted soil (CL ED73, Einheitserde, Germany) in the greenhouse, and watered with 4.7 L to field capacity. Evaporation from the soil was minimized, and no further watering was applied. Soil water content (SWC) was tracked through gravimetric measurements. The aboveground biomass of the plants (attained with the initially supplied amount of water) was determined when all plants had stopped growing and started senescing. The plant material was dried for 2 weeks at 60°C. WUEplant was calculated by dividing aboveground dry biomass by the amount of water consumed during the greenhouse experiment.
Gas exchange measurements
Measurements of gas exchange parameters were performed with an LI-6800 portable photosynthesis system (LI-COR Inc., Lincoln, NE, USA) on the youngest fully expanded leaf at plant stage V5/6. Measurements were performed at a CO2 concentration of 400 ppm, photosynthetic photon flux density of 1,500 µmol m−2s−1, leaf temperature of 25°C–26°C, and RH of 50%. The measurements were recorded every 30 s for 30–45 min. The values for A, gs and Ci in the stable phase of the recording of 10 measurements were used to calculate the respective mean values. Intrinsic WUE was calculated as A/gs. A/Ci curves were recorded at a PAR of 1,500 μmol m−2 s−1 and ambient CO2 concentrations of 635 ppm, 405 ppm, 210 ppm, 125 ppm, 90 ppm, 60 ppm, 35 ppm, and 25 ppm using an LI-6400 portable photosynthesis system (LI-COR Inc., Lincoln, NE, USA). Specific leaf area (ratio of leaf area to dry weight) was assessed on the leaf piece used to measure A/Ci curves and did not differ between RP and NIL B.
To allow the plants to recover from any disturbing impact of the gas exchange measurement, the respective leaves were harvested earliest 1 day later and snap-frozen for further analyses.
Δ13C analysis
To analyze dry matter carbon isotopic composition (δ13C), finely ground dried plant material was used. To analyze δ13C of leaves, the midvein was removed before grinding. To analyze kernels, a minimum of 5 kernels was pooled. Drying was performed either at 60°C in the oven or by lyophilization, and 3 mg of ground material was weighed into a tin capsule (5 × 8 mm IVA Analysentechnik e.K., Meerbusch, Germany) with 4 capsules per plant. Carbon isotopic composition was determined by isotope ratio mass spectrometry (Werner and Rossmann, 2015) by Isolab GmbH, Schweitenkirchen, Germany and expressed relative to the standard V-PDB.
Dry matter-derived Δ13C was calculated as described (Farquhar et al., 1989):
where δ13Cp is δ13C of the plant sample and δ13Cair is δ13C of the CO2 in the air, taken as −8‰ for all experiments.
Concurrent measurements of photosynthetic Δ13C and gas exchange
The plants were grown in a growth chamber as described above and supplied with CO2 of a controlled concentration of 400 ppm and known carbon isotopic composition mixed from dry air CO2 and tank CO2. Gas exchange measurements were performed when plants reached developmental stage V5/6 on the youngest fully developed leaf with an LI-6400 portable photosynthesis system (LI-COR Inc., Lincoln, NE, USA). To measure online Δ13C, the leaf cuvette of the LI-6400 was coupled to a continuous-flow isotope ratio mass spectrometer (IRMS, Deltaplus Advantage equipped with GasBench II, ThermoFinnigan, Bremen, Germany) as described by Gong et al. (2015). Precision (SD) of the IRMS was 0.12‰. In the leaf cuvette, a temperature of 25°C, light intensity of 1,500 µmol m−2s−1, RH of 60%, and CO2 concentration of 400 ppm were maintained for the measurements. CO2 of the same isotopic composition was supplied to the gas exchange measurement system and the growth chambers.
Δ13C was calculated as:
where ξ = Cin/(Cin – Ca), Cin is the CO2 concentration and δin is the carbon isotopic composition of the air supplied to the leaf cuvette, and Ca is the CO2 concentration and δout is the carbon isotopic composition of the air exiting the leaf cuvette. ξ was kept below 7 during measurements.
Yeast assay for ABA 8′-hydroxylase functionality
The yeast assay to detect ABA 8′-hydroxylase function was performed as described (Eggels et al., 2018) with slight modifications. Transformed yeast cells were cultivated in selective minimal medium (synthetic complete medium lacking uracil, SC-U) throughout the experiments.
Measurements of ABA and its catabolites
The contents of ABA and its catabolite in leaves were determined as previously described (Chaudhary et al., 2020). Ground leaf material (100–200 mg fresh weight), spiked with internal standard ((+)cis,trans-abscisic acid-d6 in acetonitrile), was extracted with ethyl acetate (shaking 3 × 20 s at 6,000 rpm with 40 s breaks in a bead beater; Precellys Homogenizer, Bertin Technologies, Montigny-le-Bretonneux, France). The membrane-filtered supernatant was evaporated to dryness and dissolved in acetonitrile (70 µL), before injection (2 µL) into the LC-MS/MS system.
Similarly, the PA content in the supernatant of transformed yeast was determined by spiking the sample with internal standard and directly injecting the membrane-filtrate (2 µL) into the LC-MS/MS system.
Chromatographic separation and metabolite detection were conducted by UHPLC-MS/MS. Multiple reaction monitoring transitions in positive mode for specific product ions were 263→153 (quantifier) and 263→219 (qualifier) for (+)cis,trans-abscisic acid-d0, 269→159 (quantifier) and 269→225 (qualifier) for (+)cis,trans-abscisic acid-d6, and 279→139 (quantifier) and 279→205 (qualifier) for PA-d0. ABA content is given relative to fresh weight of the leaf material.
Determination of RUBISCO and PEPC activities
Enzyme activities were measured at the Max Planck Institute for Molecular Plant Physiology (Golm, Germany). Measurement of initial and total Rubisco activities (Sulpice et al., 2007) as well as PEPC activity (Gibon et al., 2004) were based on the described protocols.
Determination of leaf chlorophyll content
Photosynthetic pigments were extracted by repeated extraction of ∼30 mg frozen, ground plant material with 1 mL 95% ethanol containing traces of CaCO3 in a Tissue Lyser II (Retsch, Qiagen, Netherlands). Absorptions of the supernatant were determined in an Epoch Microplate Spectrophotometer (BioTek, USA) at wavelengths of 649 and 664 nm and used to calculate the concentrations of chlorophyll a and b as described (Lichtenthaler, 1987).
Analysis of ABA signaling in protoplasts
Arabidopsis Col-0 protoplasts were isolated and analyzed as described (Moes et al., 2008). Approximately 105 protoplasts were transfected with 5 µg reporter plasmid (pRD29B:LUC), 3 µg control plasmid (p35S:GUS), and 5 µg DNA consisting of control and effector plasmids in varying concentrations to titrate the effector plasmid at a constant total DNA content. Effector plasmids were cloned as described (Chiasson et al., 2019), expressing the gene of interest under the control of the p35S+S1 promoter. Transformed protoplasts were incubated overnight in buffer with or without 10 µM ABA. Analyses were conducted with 6 transfections of 105 protoplasts each. Normalization of the ABA-signaling determined via luciferase activity was done by assessing GUS activity. Data are presented relative to the normalized activity using 0 µg effector plasmid incubated without ABA.
Assessment of agronomic traits
Plant height was measured from the soil to the tip of the highest leaf, after all leaves were pulled upwards.
Germination tests were conducted in three replications with 50 kernels each. After 5 min imbibition in deionized water, the kernels were placed embryo-side down between two layers of filter paper in a 20 × 20 cm2 Petri dish. The filter paper was soaked in 15 mL tap water, and an additional 5 mL of water was added after the first day of incubation at 28°C. Germinated kernels, defined based on radicle emergence, were counted on the third day, and germination rate was calculated as germinated kernels/total kernel number tested.
Thousand kernel weight was determined after manual pollination in the greenhouse. Cobs with kernel numbers <30 were excluded from analysis.
Transcript level measurements
RNA was extracted from leaves using a modified protocol (Logemann et al., 1987), followed by DNase digestion and first-strand cDNA synthesis (Maxima H Minus Kit, random hexamer primers, Thermo Scientific K1652). Reverse-transcription quantitative PCR to measure transcript levels was conducted on five leaves of individual plants, each with three qPCR reactions. The reaction was performed using KAPA SYBR FAST (Thermo Fisher Scientific, USA) in a QuantStudio™ 3 system (Thermo Fisher Scientific, USA). In the ZmAbh4 gene, primers were located in the exons, spanning each intron (Supplemental Table S4). Data acquisition was conducted as described by the supplier, with the respective annealing temperature for each primer pair (Supplemental Table S4) and 58°C for the housekeeping gene (ZmMep). The transcript quantities of each replicate were calculated relative to ZmMep in the sample (primers: 5′-TGTACTCGGCAATGCTCTTG-3′, 5′- TTTGATGCTCCAGGCTTACC-3′).
Detection of the 18 kb intron insertion in a panel of 33 maize lines
The tested set included maize inbred lines from four different heterotic groups: Stiff Stalk dent (B73, EC169, F618, F98902, D06, D09, UH250), non-Stiff Stalk dent (RP, PH207, F353, UH304, F252, Mo17 and W117), Northern flint (DP, CH10, DK105, D152, FF0721H-7, UH006, UH007, UH009, F283, F2, F7, F03802, EP1), and non-Northern Flint (Lo11, EC49A, EP44, F64, EZ5), as well as Teosinte.
Two PCRs were performed using a common forward primer (5′-GCACACGAAGAGCATG-3′) located in exon 4 of the ZmAbh4 gene and two alternative reverse primers located in the 4th intron either within (5′-CTGTGTTCTTTAGAGATATACTT-3′) or outside (5′-CTCTCGTTGCTCCT-3′) the 18 kb insertion. The reactions were performed using Q5® Hot Start High-Fidelity DNA Polymerase (New England Biolabs Inc., Ipswich, MA, USA) according to the supplier with annealing at 60°C. Amplification of respective gene fragments was verified by Sanger sequencing of the amplicons after separation by agarose gel electrophoresis and purification (NucleoSpin® Gel and PCR Clean-up kit, Macherey-Nagel GmbH & Co. KG, Düren, Germany).
Sanger sequencing
Sanger sequencing was performed using Mix2seq kits at Eurofins Genomics Germany GmbH (Ebersberg, Germany).
PacBio Long reads
High Molecular Weight DNA isolation and preparation of PacBio CLR libraries with insert size above 20 kb were performed in CNRGV, INRA Occitanie Toulouse, France (cnrgv.toulouse.inra.fr). PacBio long reads of the RP and NIL B genomes were obtained from massive parallel sequencing performed at the National Genomics Infrastructure (NGI)/Uppsala Genome Center, Uppsala, Sweden.
Statistical analyses
Analyses were conducted in R Studio (RStudio Team, 2020). For significance testing, two-sided Student’s t tests were conducted between RP and NIL B. When NILs were compared to the common reference genotype RP, two-sided Dunnett’s test was used (R package “multcomp”; Hothorn et al., 2008). For comparisons among all genotypes, LSD with correction for multiple testing was applied. One-sided Student’s t tests were used for significance testing between mutants and WTs (Supplemental Data Set S2).
Accession numbers
The sequences of genes used in this study are listed in NCBI under the following accession numbers:
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. RP and NIL B differ in carbon isotope discrimination measured in kernels (Δ13CKernel), water use efficiency (WUE), gas exchange parameters and levels of abscisic acid (ABA).
Supplemental Figure S2. Genetic composition of maize near-isogenic lines NIL B, G_1 and G_2, and recurrent parent RP.
Supplemental Figure S3. Transcript levels of ZmAbh4 in recurrent parent RP and near-isogenic line NIL B, measured by quantitative RT-PCR with primers spanning the different introns.
Supplemental Figure S4 . CRISPR/Cas9 mutants show loss of function of ZmAbh4.
Supplemental Figure S5. Agronomic traits of NILs and CRISPR/Cas9 mutants.
Supplemental Figure S6. Measurement of carbon isotope discrimination (Δ13C) and its connection to intrinsic water use efficiency (iWUE).
Supplemental Figure S7. Photosynthesis-related traits in RP and NIL B.
Supplemental Figure S8. Gel electrophoresis detecting the presence or absence of the 18kb insertion in the fourth intron of the ZmAbh4 gene (Zm00001d020717; encoding ABA 8′-hydroxylase 4) in 33 maize inbred lines.
Supplemental Table S1. Regions with genomic introgressions from a donor parent in the background of a recurrent parent in ten near-isogenic lines (NILs).
Supplemental Table S2. Phenotyping of near-isogenic maize lines (NILs) in a greenhouse and a growth chamber experiment.
Supplemental Table S3. Sequence comparisons between NIL B and RP alleles of ZmAbh4.
Supplemental Table S4. Primers used for the qPCR analysis described in Supplemental Figure S3.
Supplemental Data Set S1. Complete genomic sequences of the NIL B and RP ZmaAbh4 alleles.
Supplemental Data Set S2. Statistical analyses.
Supplementary Material
Acknowledgements
We thank Sylwia Schepella, Stefan Schwertfirm, Brigitte Neuhauser, Margot Siebler, Iris Leineweber, and Georg Maier for excellent technical support. The transformation to generate CRISPR/Cas9 mutants was performed at the VIB Center for Plant Systems Biology, Ghent, Belgium under the supervision of Laurens Pauwels, who also helped with construct design. We thank the Plant Technology Center (Technical University of Munich, Germany) for providing infrastructure and technical support during greenhouse and growth chamber experiments. We thank Michael Gigl and Wilfried Rozhon for ABA and PA measurements. We thank Thorsten E. E. Grams for critical discussions. We further acknowledge Simon Mayer for support in the drought stress experiment and Shen Yeng Tan for gene expression measurements. We would like to acknowledge the support of Sandrine Arribat and Caroline Callot from CNRGV, INRA and the National Genomic Infrastructure (NGI)/Uppsala Genome Center for providing assistance in massive parallel sequencing and computational infrastructure.
Funding
This study was funded by the German Research Foundation (Deutsche Forschungsgemeinschaft; DFG) through the Sonderforschungsbereich 924 (SFB924): “Molecular mechanisms regulating yield and yield stability in plants”, and by the Federal Ministry of Education and Research (BMBF, Germany) through the project “Maximizing photosynthetic efficiency in maize (FullThrottle)” within the scope of the funding initiative “Plant Breeding Research for the Bioeconomy” (Funding ID: 031B0205A and 031B0205C). Work performed at NGI/Uppsala Genome Center has been funded by RFI/VR and Science for Life Laboratory, Sweden.
Conflict of interest statement. None declared.
Contributor Information
Sonja Blankenagel, Plant Breeding, TUM School of Life Sciences, Technical University of Munich, Liesel-Beckmann-Straße 2, 85354 Freising, Germany.
Stella Eggels, Plant Breeding, TUM School of Life Sciences, Technical University of Munich, Liesel-Beckmann-Straße 2, 85354 Freising, Germany.
Monika Frey, Plant Breeding, TUM School of Life Sciences, Technical University of Munich, Liesel-Beckmann-Straße 2, 85354 Freising, Germany.
Erwin Grill, Botany, TUM School of Life Sciences, Technical University of Munich, Emil-Ramann-Straße 2, 85354 Freising, Germany.
Eva Bauer, Campus Office, TUM School of Life Sciences, Technical University of Munich, Weihenstephaner Steig 22, 85354 Freising, Germany.
Corinna Dawid, Food Chemistry and Molecular Sensory Science, TUM School of Life Sciences, Technical University of Munich, Lise-Meitner-Straße 34, 85354 Freising, Germany.
Alisdair R Fernie, Central Metabolism, Max Planck Institute of Molecular Plant Physiology, Am Mühlenberg 1, 14476 Potsdam, Germany.
Georg Haberer, Plant Genome and Systems Biology, Helmholtz Center Munich, Ingolstädter Landstr. 1, 85764 Neuherberg, Germany.
Richard Hammerl, Food Chemistry and Molecular Sensory Science, TUM School of Life Sciences, Technical University of Munich, Lise-Meitner-Straße 34, 85354 Freising, Germany.
David Barbosa Medeiros, Central Metabolism, Max Planck Institute of Molecular Plant Physiology, Am Mühlenberg 1, 14476 Potsdam, Germany.
Milena Ouzunova, KWS SAAT SE, Grimsehlstraße 31, 37555 Einbeck, Germany.
Thomas Presterl, KWS SAAT SE, Grimsehlstraße 31, 37555 Einbeck, Germany.
Victoria Ruß, Botany, TUM School of Life Sciences, Technical University of Munich, Emil-Ramann-Straße 2, 85354 Freising, Germany.
Rudi Schäufele, Grassland, TUM School of Life Sciences, Technical University of Munich, Alte Akademie 12, 85654 Freising, Germany.
Urte Schlüter, Institute of Plant Biochemistry, Cluster of Excellence on Plant Science (CEPLAS), Heinrich Heine University Düsseldorf, Universitätsstraße 1, 40225 Düsseldorf, Germany.
Francois Tardieu, Université de Montpellier, INRAE, Laboratoire d’Ecophysiologie des Plantes sous Stress Environnementaux (LEPSE), Place Viala, F-34060, Montpellier, France.
Claude Urbany, KWS SAAT SE, Grimsehlstraße 31, 37555 Einbeck, Germany.
Sebastian Urzinger, Plant Breeding, TUM School of Life Sciences, Technical University of Munich, Liesel-Beckmann-Straße 2, 85354 Freising, Germany.
Andreas P M Weber, Institute of Plant Biochemistry, Cluster of Excellence on Plant Science (CEPLAS), Heinrich Heine University Düsseldorf, Universitätsstraße 1, 40225 Düsseldorf, Germany.
Chris-Carolin Schön, Plant Breeding, TUM School of Life Sciences, Technical University of Munich, Liesel-Beckmann-Straße 2, 85354 Freising, Germany.
Viktoriya Avramova, Plant Breeding, TUM School of Life Sciences, Technical University of Munich, Liesel-Beckmann-Straße 2, 85354 Freising, Germany.
V.A., S.B., S.E., and M.F. designed and performed the experiments and analyzed the data; R.H and C.D. were responsible for the abscisic and phaseic acid quantification; G.H. and S.U. analyzed the PacBio long reads; D.B.M. was responsible for the enzyme measurements; V.R. contributed to the analysis of abscisic acid signaling in protoplasts; R.S. contributed to the online carbon isotope discrimination measurements; E.B., M.O., T.P., and C.U. developed the plant material; C.-C.S., V.A., M.F., E.B., A.P.M.W., E.G., U.S., F.T., R.S., M.O., and A.R.F. designed the research and developed ideas; S.E., S.B., and V.A. wrote the manuscript, C.-C.S., M.F., E.G., and F.T. edited the manuscript; all authors read and approved the final manuscript; V.A. agrees to serve as the author responsible for contact and to ensure communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Viktoriya Avramova (viktoriya.avramova@tum.de).
References
- Avramova V, Meziane A, Bauer E, Blankenagel S, Eggels S, Gresset S, Grill E, Niculaes C, Ouzunova M, Poppenberger B, et al. (2019) Carbon isotope composition, water use efficiency, and drought sensitivity are controlled by a common genomic segment in maize. Theor Appl Genet 132: 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino LT, Caine RS, Gray JE (2019) Impact of stomatal density and morphology on water-use efficiency in a changing world. Front Plant Sci 10: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugière N, Zhang W, Xu Q, Scolaro EJ, Lu C, Kahsay RY, Kise R, Trecker L, Williams RW, Hakimi S, et al. (2017) Overexpression of RING domain E3 ligase ZmXerico1 confers drought tolerance through regulation of ABA homeostasis. Plant Physiol 175: 1350–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary A, Chen X, Gao J, Lesniewska B, Hammerl R, Dawid C, Schneitz K (2020) The Arabidopsis receptor kinase STRUBBELIG regulates the response to cellulose deficiency. Plos Genet 16: e1008433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chang SX, Anyia AO (2011) Gene discovery in cereals through quantitative trait loci and expression analysis in water-use efficiency measured by carbon isotope discrimination. Plant Cell Environ 34: 2009–2023 [DOI] [PubMed] [Google Scholar]
- Chiasson D, Gimenez-Oya V, Bircheneder M, Bachmaier S, Studtrucker T, Ryan J, Sollweck K, Leonhardt H, Boshart M, Dietrich P, et al. (2019) A unified multi-kingdom Golden Gate cloning platform. Sci Rep 9: 10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet JP, Haeussler M (2018) CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res 46: W242–W245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon AG, Richards RA, Rebetzke GJ, Farquhar GD (2004) Breeding for high water-use efficiency. J Exp Bot 55: 2447–2460 [DOI] [PubMed] [Google Scholar]
- Eggels S, Blankenagel S, Schön CC, Avramova V (2021) The carbon isotopic signature of C4 crops and its applicability in breeding for climate resilience. Theor Appl Genet 134: 1663–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggels S, Avramova V, Schön CC, Poppenberger B, Rozhon W (2018) Assay for abscisic acid 8'-hydroxylase activity of cloned plant cytochrome P450 oxidases in Saccharomyces cerevisiae. Anal Biochem 553: 24–27 [DOI] [PubMed] [Google Scholar]
- Ellsworth PZ, Feldman MJ, Baxter I, Cousins AB (2020) A genetic link between leaf carbon isotope composition and whole-plant water use efficiency in the C4 grass Setaria. Plant J 102: 1234–1248 [DOI] [PubMed] [Google Scholar]
- Farquhar GD (1983) On the nature of carbon isotope discrimination in C4 species. Funct Plant Biol 10: 205–226 [Google Scholar]
- Farquhar GD, Ehleringer JR, Hubick KT (1989) Carbon isotope discrimination and photosynthesis. Annu Rev Plant Phys 40: 503–537 [Google Scholar]
- Feys K, Demuynck K, De Block J, Bisht A, De Vliegher A, Inze D, Nelissen H (2018) Growth rate rather than growth duration drives growth heterosis in maize B104 hybrids. Plant Cell Environ 41: 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hohne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M (2004) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong XY, Schaufele R, Feneis W, Schnyder H (2015) 13CO2/12CO2 exchange fluxes in a clamp-on leaf cuvette: disentangling artefacts and flux components. Plant Cell Environ 38: 2417–2432 [DOI] [PubMed] [Google Scholar]
- Gresset S, Westermeier P, Rademacher S, Ouzunova M, Presterl T, Westhoff P, Schön CC (2014) Stable carbon isotope discrimination is under genetic control in the C4 species maize with several genomic regions influencing trait expression. Plant Physiol 164: 131–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD (2004) Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429: 268–274 [DOI] [PubMed] [Google Scholar]
- Henderson S, von Caemmerer S, Farquhar GD, Wade LJ, Hammer G (1998) Correlation between carbon isotope discrimination and transpiration efficiency in lines of the C4 species Sorghum bicolor in the glasshouse and the field. Aust J Plant Physiol 25: 111–123 [Google Scholar]
- Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biometrical J 50: 346–363 [DOI] [PubMed] [Google Scholar]
- Hufford MB, Seetharam AS, Woodhouse MR, Chougule KM, Ou SJ, Liu JN, Ricci WA, Guo TT, Olson A, Qiu YJ, et al. (2021) De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science 373: 655–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao YP, Peluso P, Shi JH, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei XH, Chin CS, et al. (2017) Improved maize reference genome with single-molecule technologies. Nature 546: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krochko JE, Abrams GD, Loewen MK, Abrams SR, Cutler AJ (1998) (+)-abscisic acid 8 '-hydroxylase is a cytochrome P450 monooxygenase. Plant Physiology 118: 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leakey ADB, Ferguson JN, Pignon CP, Wu A, Jin ZN, Hammer GL, Lobell DB (2019) Water use efficiency as a constraint and target for improving the resilience and productivity of C3 and C4 crops. Annu Rev Plant Biol 70: 781–808 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK (1987) Chlorophylls and carotenoids - pigments of photosynthetic biomembranes. Method Enzymol 148: 350–382 [Google Scholar]
- Liu SX, Li CP, Wang HW, Wang SH, Yang SP, Liu XH, Yan JB, Li BL, Beatty M, Zastrow-Hayes G, et al. (2020) Mapping regulatory variants controlling gene expression in drought response and tolerance in maize. Genome Biol 21: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant-tissues. Anal Biochem 163: 16–20 [DOI] [PubMed] [Google Scholar]
- Mega R, Abe F, Kim JS, Tsuboi Y, Tanaka K, Kobayashi H, Sakata Y, Hanada K, Tsujimoto H, Kikuchi J, et al. (2019) Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat Plants 5: 153–159 [DOI] [PubMed] [Google Scholar]
- Mittelheuser CJ, Van Steveninck RFM (1969) Stomatal closure and inhibition of transpiration induced by (RS)-abscisic acid. Nature 221: 281–282 [Google Scholar]
- Moes D, Himmelbach A, Korte A, Haberer G, Grill E (2008) Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J 54: 806–819 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Tanaka Y, Abrams SR, Kamiya Y, Seki M, Nambara E (2009) High humidity induces abscisic acid 8'-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis. Plant Physiol 149: 825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, Lugo CSB, Elliott TA, Ware D, Peterson T, et al. (2019) Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol 20: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portwood JL 2nd, Woodhouse MR, Cannon EK, Gardiner JM, Harper LC, Schaeffer ML, Walsh JR, Sen TZ, Cho KT, Schott DA, et al. (2019) MaizeGDB 2018: the maize multi-genome genetics and genomics database. Nucleic Acids Res 47: D1146–D1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah SK, Reddy KR, Li J (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7: 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Tschoep H, VON Korff M, Bussis D, Usadel B, Hohne M, Witucka-Wall H, Altmann T, Stitt M, Gibon Y (2007) Description and applications of a rapid and sensitive non-radioactive microplate-based assay for maximum and initial activity of D-ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Cell Environ 30: 1163–1175 [DOI] [PubMed] [Google Scholar]
- Tardieu F, Simonneau T, Muller B (2018) The physiological basis of drought tolerance in crop plants: a scenario-dependent probabilistic approach. Annu Rev Plant Biol 69: 733–759 [DOI] [PubMed] [Google Scholar]
- Twohey RJ 3rd, Roberts LM, Studer AJ (2019) Leaf stable carbon isotope composition reflects transpiration efficiency in Zea mays. Plant J 97: 475–484 [DOI] [PubMed] [Google Scholar]
- Unterseer S, Bauer E, Haberer G, Seidel M, Knaak C, Ouzunova M, Meitinger T, Strom TM, Fries R, Pausch H, et al. (2014) A powerful tool for genome analysis in maize: development and evaluation of the high density 600 k SNP genotyping array. BMC Genomics 15: 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhaneni R, Wurtzel ET (2010) From epoxycarotenoids to ABA: the role of ABA 8'-hydroxylases in drought-stressed maize roots. Arch Biochem Biophys 504: 112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venzhik Y, Talanova V, Titov A (2016) The effect of abscisic acid on cold tolerance and chloroplasts ultrastructure in wheat under optimal and cold stress conditions. Acta Physiol Plant 38: 63 [Google Scholar]
- von Caemmerer S, Ghannoum O, Pengelly JJ, Cousins AB (2014) Carbon isotope discrimination as a tool to explore C4 photosynthesis. J Exp Bot 65: 3459–3470 [DOI] [PubMed] [Google Scholar]
- Wang HB, Zhao S, Mao K, Dong QL, Liang BW, Li C, Wei ZW, Li MJ, Ma FW (2018) Mapping QTLs for water-use efficiency reveals the potential candidate genes involved in regulating the trait in apple under drought stress. Bmc Plant Biol 18: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner RA, Rossmann A (2015) Multi element (C, H, O) stable isotope analysis for the authentication of balsamic vinegars. Isotopes Environ Health Stud 51: 58–67 [DOI] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. Bmc Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZY, Liu JH, Tischer SV, Christmann A, Windisch W, Schnyder H, Grill E (2016) Leveraging abscisic acid receptors for efficient water use in Arabidopsis. Proc Natl Acad Sci USA 113: 6791–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZY, Liu JH, Poree F, Schaeufele R, Helmke H, Frackenpohl J, Lehr S, von Koskull-Doring P, Christmann A, Schnyder H, Schmidhalter U, Grill E (2019) Abscisic acid receptors and coreceptors modulate plant water use efficiency and water productivity. Plant Physiol 180: 1066–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DP (2014) Abscisic Acid: Metabolism, Transport and Signaling. Springer, Dordrecht, The Netherlands [Google Scholar]
- Zhang HM, Li GL, Fu C, Duan SN, Hu D, Guo XL (2020) Genome-wide identification, transcriptome analysis and alternative splicing events of Hsf family genes in maize. Sci Rep-Uk 10: 8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.