Figure 1.
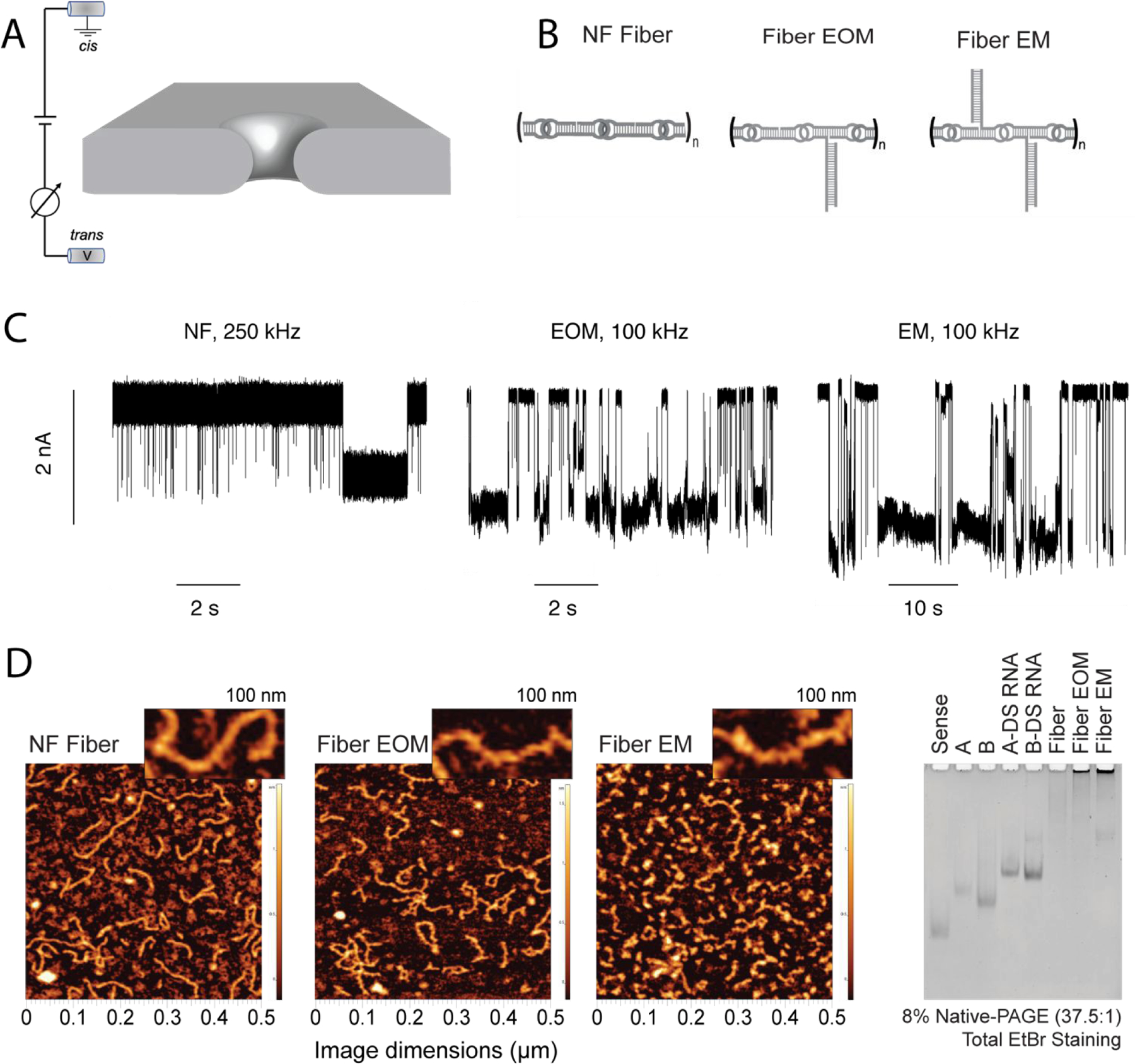
Differentiation of RNA fibers using a solid-state nanopore. (A) Schematic representation of experimental set-up, wherein the application of positive transmembrane voltages electrophoretically captures NANPs at the pore vicinity. (B) Sketches of the different RNA fibers studied in this work: non-functionalized (NF) fibers; fibers with a branch at every other monomer (EOM); fibers with a branch at every monomer (EM). (C) Representative fragments of ionic current traces obtained using a 4.5 nm diameter nanopore at 300 mV (in 1 M KCl, 10 mM HEPES, 2 mM MgCl2, pH 7.5) for NF, EOM, and EM fibers. The traces from the NF fiber sample were recorded at a sampling rate of 4,167 kHz and low-pass filtered at 250 kHz, whereas traces from the EOR and EM fiber samples (concentration of 25–100 nM) were recorded at a sampling rate of 250 kHz and low-pass filtered at 100 kHz. These traces clearly demonstrate that the fibers can be identified by solid-state nanopore according to their branching, which is difficult to achieve using other methods such as AFM and gel electrophoresis as shown in (D). (D) AFM images and gel electrophoresis of each fiber, suggesting large heterogeneity in the fibers lengths.
