Figure 2. Advancements in Base Excision Repair.
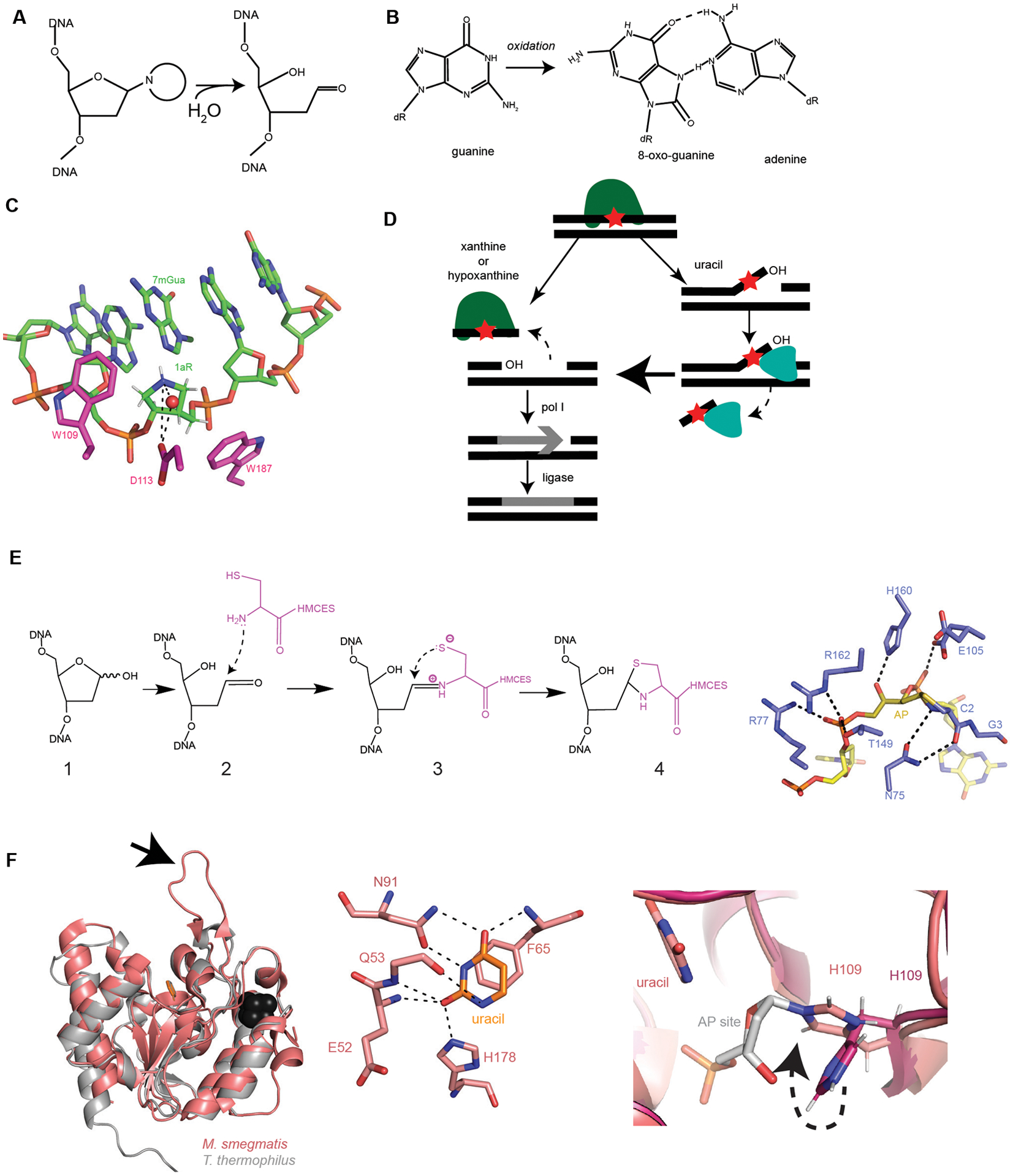
(A) Formation of an AP site through hydrolysis of the N-glycosidic bond (adapted from129). (B) Guanine oxidation into 8-oxo-guanine and mismatched pairing to adenine. (C) AlkD (5kub, shown in pink) protein contacts with depurination intermediate analogue 1aR (1-aza-2’, 4’-dideoxyribose) within DNA (shown in green). Red sphere is a water nucleophile. Dashed black lines are hydrogen bonds. CH/π interactions with W109 and W187 to 1aR (adapted from52). (D) Summary of the EndoV (YwqL)/ExoA repair of deaminated bases (adapted from63). EndoV identifies and incises 3’ of the lesion. If the lesion is xanthine or hypoxanthine, EndoV cuts 5’ of the lesion and pol I fills in the gap. DNA ligase seals the nick to complete repair. If the lesion is uracil or an AP site, the initial endonuclease activity creates a 3’ flap containing the lesion. ExoA uses its 3’ to 5’ exonuclease activity to remove the lesion containing strand, followed by pol I and ligase. (E) Left: Proposed stepwise formation of the thiazolidine linkage between HMCES and DNA based on YedK. 1. AP site within DNA, 2. Nucleophilic attack on AP site, 3. Schiff base intermediate, 4. Thiazolidine link between HMCES and DNA. Right: YedK (6nua, shown in purple) DNA-protein crosslinks with AP site (dark yellow) within DNA (yellow). Critical hydrogen bonds between protein and DNA shown as black dashed lines (adapted from56). Note: E105 is shown with two conformers. (F) Left: Mycobacterium tuberculosis UdgX in salmon pink (6ioa) aligned with a family 4 UDG from Thermus thermophilus in light gray (1ui0), uracil shown in orange and Fe-S cluster in black. Arrow pointing at characteristic protruding loop (adapted from61). Middle: Mycobacterium tuberculosis UdgX (6ioa) interactions with uracil (orange). H178 is the residue responsible for excision of uracil. H178, along with E52, Q53, F65, and N91 hydrogen bond to uracil. Black dashed lines are hydrogen bonds (adapted from60). Right: Mycobacterium tuberculosis apo-UdgX (6ail) in magenta aligned with uracil-bound in salmon (6ajo). H109 changes position within the enzyme (shown with dashed black arrow) when uracil is bound and excised from the DNA backbone to form a covalent bond with the AP site (in gray). H109 serves as the nucleophile instead of water.
