Figure 6. MrfA involvement in DNA repair.
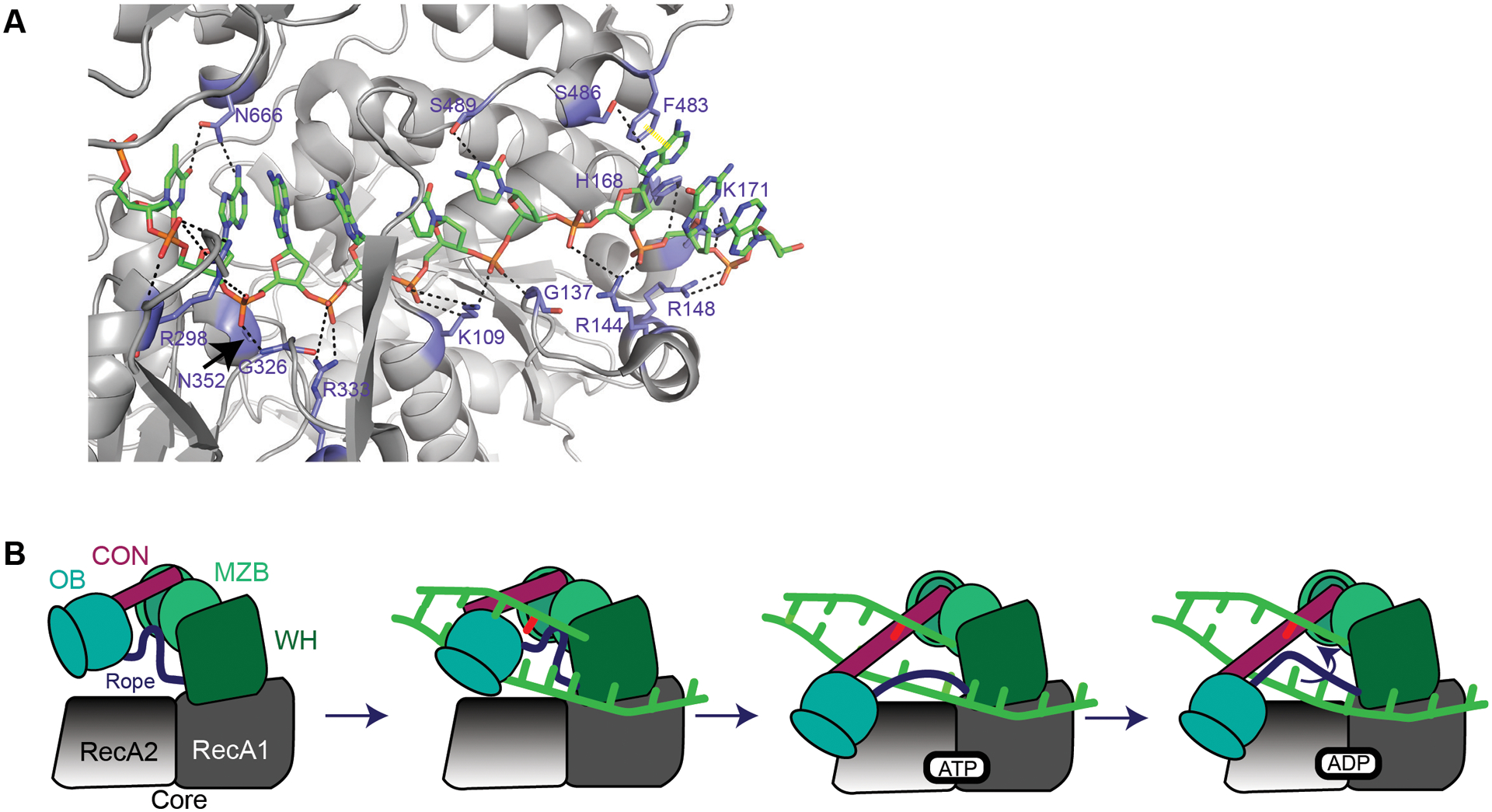
(A) MrfA (6znq) contacts with DNA in the loading strand adapted from128. Note: K108128 is K109. Black dashed lines represent hydrogen bonds. Yellow dashed line represents π-π stacking. A and B are adapted from the following work128. (B) It is unknown how MrfA loads onto DNA or how it identifies a mitomycin C-induced lesion. MrfA contains seven domains: a N-terminal region (NTR, not shown), RecA1 and RecA2 (core domains, grey), a winged helix domain (WHD, green), an oligonucleotide/oligosaccharide binding domain (OB, blue), a connector element (CON, maroon), and a C-terminal DUF1998 that contains a unique MrfA zinc binding domain (MZB, light green)128. DNA shown in bright green with a DNA lesion in red. Upon loading DNA, the OB domain shifts toward RecA2 domain128. The OB domain binds RecA2. As a result, the rope (purple) connecting the OB domain and winged-helix domain undergoes a conformational change. The tightening of this rope across the ssDNA prevents the loading strand from slipping backward128. ATP is bound, causing DNA translocation through an “inchworm”-like mechanism. The mechanism for coupling DNA translocation and unwinding is unknown128. As new ATP is hydrolyzed, DNA is pumped through the protein and products are released.
