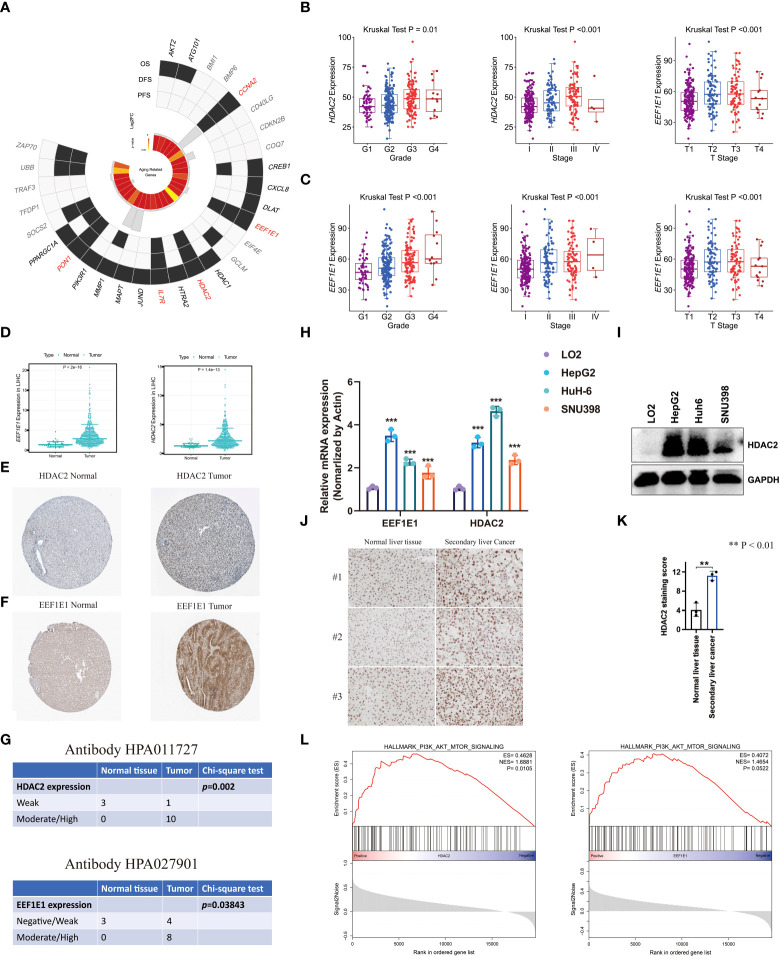
Figure 8.
Identification of aging-related gene hubs associated with liver cancer disease progression. (A) Five aging-related genes were selected by using Kaplan–Meier estimates of overall survival, progression-free survival, and disease-free survival. (B, C) Two aging-related gene hubs were selected by using the correlation between aging-related genes and clinical feature. (D) The expression level of HDAC2 and EEF1E1 in adjacent normal liver and LIHC samples. (E, F) The protein expression level of HDAC2 and EEF1E1 using the HPA database. (G) Chi-square test of the significance between the number of normal tissue samples and hepatocellular carcinoma samples in protein expression level of HDAC2 and EEFIE1. (H) qPCR analysis of EEF1E1 or HDAC2 expression in the hepatocyte line LO2 and hepatocellular carcinoma cell lines HepG2, Huh6, and SNU398. The experiment was performed three times with consistent results; representative results from one set of experiments are shown. **P < 0.01; ***P < 0.001.(I) Immunoblot analysis of HDAC2 levels in the hepatocyte line LO2 and hepatocellular carcinoma cell lines HepG2, Huh6, and SNU398. Experiment was performed three times with consistent results; representative image is shown. (J) IHC analysis of HDAC2 in normal liver tissues and secondary liver cancer tissues. (K) HDAC2 staining score in normal liver tissues and secondary liver cancer tissues. (L) GSEA was employed to explore the biological function of these aging-related genes (HDAC2 and EEF1E1) in the progression of LIHC.