Abstract
The aim of the present study was to decipher the mechanism of glutathione-S-transferase Ω-1 (GSTO1)-induced drug resistance in colon cancer cells. Cisplatin is used widely as a therapeutic drug in cancer, but colon cancer is the most susceptible to acquired drug resistance. Autophagy is recognized as one of the contributors to drug resistance in cancers. Phase II detoxifying enzymes, such as GSTO1, serve important roles in autophagy-apoptosis cross talk. The present study revealed a novel interaction between GSTO1 and TNFα-induced protein 3/zinc-finger protein A20 (TNFαIP3/A20) as a prime target for cisplatin sensitization in drug-resistant cells. GSTO1 and ATP-binding cassette subfamily B member 1 (ABCB1) were both expressed at higher levels in multidrug-resistant (MDR) HCT-116 cells compared with the wild-type (WT) HCT-116 cells, suggesting they may serve vital roles in multidrug resistance. MDR cells showed autophagy induction, which is dependent on calcium signaling-dependent endoplasmic stress. In WT cells, the mitochondria-dependent pathway leads to apoptosis, which was not observed in MDR cells. The MDR conditions were mimicked by transfecting WT cells with the GSTO1-activation CRISPR plasmid, which induced autophagy. Similarly, MDR cells with GSTO1-knockdown (KD) CRISPR/Cas9 transfection showed reduced autophagy with increased apoptosis. These data revealed a potentially important role of GSTO1 in drug resistance. A GSTO1 pull-down assay detected TNFαIP3/A20 as a binding partner in MDR cells. The data suggested that the expression of TNFαIP3/A20 may be dependent on GSTO1 expression in MDR cells. Targeting either GSTO1 or TNFαIP3/A20 by CRISPR/Cas9 sensitized the MDR cells to cisplatin. GSTO1 and TNFαIP3/A20 dual-KD cells were more sensitive to cisplatin compared with single-gene KD cells. These data highlight the importance of the GSTO1-TNFαIP3/A20 interaction during drug resistance.
Keywords: glutathione-S-transferase Ω-1, TNFαIP3/A20, drug resistance, autophagy, apoptosis
Introduction
Multidrug resistance can lead to the failure of cancer chemotherapy and is a major cause of mortality in cancer patients (1). Cancer cells acquire resistance to drugs through various mechanisms, such as drug effluxes and xenobiotic-mediated detoxification (2). Cisplatin is one of the most frequently used drugs in a number of cancers; however, colon cancer shows strong resistance to cisplatin (3). The autophagy-mediated cell survival pathway has emerged as a prime mechanism for multidrug resistance (4), but our understanding of autophagy-mediated multidrug resistance is still lacking.
Glutathione S-transferases (GSTs) are multigene family phase-II detoxifying enzymes activated during the resistance to chemotherapeutic agents in several cancers, including colon, esophageal and breast cancer (5-7). The omega (Ω) class of GSTs (GSTOs) are unique among the GST family, having a cysteine residue in their active site instead of tyrosine or serine (8). A previous publication suggested that GSTO1 serves a key role in autophagy-apoptosis crosstalk. GSTO1 inhibits the JNK-mediated apoptosis signaling pathway in macrophages (9). A previous publication documented the upregulation of GSTO1 in chemotherapy-exposed breast cancer cells (10). The knockdown of GSTO1 in these cells reduced the formation of carboplatin-induced breast cancer stem cells and decreased tumor initiation. The authors suggested a novel interaction between GSTO1 and ryanodine receptor 1, which accelerates calcium release from the endoplasmic reticulum (ER) and eventually activates the STAT3 pathway (10). Proteome-based analysis of platinum-resistant human ovarian cancer cell lines revealed the stable upregulation of GSTO1 expression compared with the parental cells (11). Gene expression profiling of proteasome inhibitor-resistant human myeloma cell lines also revealed an upregulation of GSTO1 compared with the parental cells (12).
Previous studies have suggested that autophagy and drug resistance are causes of cancer metastasis (13-15). Research on metastasis also suggested that the incidence of autophagy increases more in metastasized tumors compared with the levels of autophagy in the original tumor (13,15). Autophagy also modulates tumor cell invasion and epithelial-to-mesenchymal transition (13). Moreover, a previous publication suggested that drug-resistant cancer cells show greater invasive ability, leading to chemotherapy failure (14). The conclusion drawn from the present study suggested that GSTO1 may control autophagy, which, in turn, may control drug resistance in colon cancer cells and may be involved in regulating metastasis.
The present study results suggested that GSTO1 may be a crucial factor between cisplatin sensitization and resistance. GSTO1 is a protein that prompts MDR cells into autophagy cell survival. In addition, TNFαIP3/A20 was shown to interact very strongly with GSTO1 in MDR cells. The GSTO1-TNFαIP3/A20 interaction is one of the mechanisms of drug resistance, and the inhibition of those mechanisms in MDR cells sensitizes them to cisplatin.
Materials and methods
Chemicals
5-Fluorouracil (5-FU), cisplatin, docetaxel, vincristine, Hoechst 33342, bafilomycin A1 (BAF-A1), dichloro-dihydrofluorescein diacetate (DCFH-DA), Fura-2/AM and monodansylcadaverine (MDC) were purchased from MilliporeSigma (Merk KGaA).
Cell culture, establishment of the MDR HCT-116 cell line and treatment with BAF-A1
The HCT-116 human colorectal carcinoma cell line (American Type Culture Collection) was maintained at 37°C and 5% CO2 in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS, 25 mM HEPES and 1% penicillin/streptomycin cocktail (Gibco; Thermo Fisher Scientific, Inc.). Drug-resistant cells were generated using a stepwise increase in treatment doses with docetaxel, vincristine, cisplatin and 5-FU following a protocol from a previously published article by our group (16). For the initial dose, HCT-116 cells were treated with 0.2 nM docetaxel, 0.2 nM vincristine, 0.5 µM cisplatin and 0.5 µM 5-FU until the cells became stable in these drug doses. Stable cells were exposed three times to the drug combination over a three-day period for 3-4 weeks, allowing growth recovery between cycles. After completing three drug treatment cycles, the doses were doubled, and the procedure was repeated until treatment with the final drug concentrations was achieved over an 8-10-month period. The MDR subline was maintained in complete RPMI-1640 medium containing final drug concentrations of 10 nM docetaxel, 10 nM vincristine and 10 µM cisplatin and 10 µM 5-FU.
To inhibit autophagy induction in MDR cells, 1×106 cells were treated with 10 nM of BAF-A1 for 24 h at 37°C and 5% CO2 in complete media; 0.1% DMSO was used as a vehicle. Cells were subsequently used for protein isolation and western blot analysis.
Protein isolation and western blot analysis
Whole-cell proteins were isolated using RIPA lysis buffer (MilliporeSigma; Merck KGaA) according to the manufacturer's protocol. The cytosolic and nuclear fractions were isolated using a NE-PER Nuclear Protein Extraction Kit (Thermo Scientific; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The mitochondrial protein was obtained by isolating the mitochondria from the cells using a Mitochondria Isolation kit (MilliporeSigma; Merck KGaA); proteins were extracted from the isolated mitochondria using a RIPA lysis buffer. Protein samples were quantified using a BCA Protein Assay Kit (Cell Signaling Technology, Inc.) according to the manufacturer's protocol.
Proteins (10 µg/lane for whole cell lysates; 20 µg/lane for nuclear lysates; and 30 µg/lane for mitochondrial lysates) were resolved by 8-15% SDS-PAGE, based on molecular weight of proteins, and transferred to a PVDF membrane (Roche Diagnostics). The membranes were then incubated overnight with primary antibodies at 4°C. The membranes were then incubated with the respective secondary antibodies for 1 h at room temperature and visualized by ECL Western Blotting detection reagent (cat. no. RPN2109; Amersham; Cytiva) according to the recommended procedure. β-actin, Lamin B1 and heat-shock protein 60 (HSP60) were used as markers for the whole cell, nucleus and mitochondria, respectively. Tables SI and SII list the primary and secondary antibodies used for western blot analysis, respectively. Densitometric analysis of the protein bands was performed using ImageJ (version 1.53f) open-source program (National Institutes of Health). β-actin, Lamin B1 and HSP60 were used as loading controls to normalize protein expression in whole-cell lysates, nuclear lysates and mitochondrial lysates, respectively.
Lactate dehydrogenase (LDH) assay
WT and MDR HCT-116 cells were seeded in a 48-well plate at 1×105 cells/well and treated with 50 µM cisplatin and vehicle for 12 h at 37°C and 5% CO2 in complete medium. The cisplatin and vehicle-treated cell culture supernatants were collected and LDH release from the cells was quantified using an LDH cytotoxicity assay kit (cat. no. TOX7; MilliporeSigma; Merck KGaA) according to the manufacturer's protocol. The primary absorbance was measured at 490 nm and background absorbance was measured at 690 nm using a microplate reader (BioTek Instruments, Inc.). Actual absorbance was calculated by subtracting background absorbance value from the primary absorbance value.
Hoechst staining for chromatin condensation
WT and MDR HCT-116 cells were seeded in a 4 well chambered slide at 1×105 cells/well and treated with 50 µM cisplatin and vehicle for 12 h at 37°C and 5% CO2 in complete medium. Briefly, the cisplatin- and vehicle-treated cells were subsequently fixed with 4% formaldehyde at room temperature, washed with PBS and incubated with Hoechst 33342 (1 µg/ml) at 37°C for 10 min. After washing with PBS, the DNA chromatin morphological features were analyzed using a Nikon Eclipse fluorescence microscope (Nikon Corporation).
Calcium measurement
WT and MDR HCT-116 cells were seeded in a 4-well chambered slide at 1×104 cells/well. Calcium levels were measured by incubating the cisplatin and vehicle-treated cells with 5 µM Fura-2/AM in Hank's balanced salt solution (HBSS) buffer for 60 min at 37°C. The samples were then washed three times with HBSS at 37°C and fluorescence micrographs were taken using a Nikon Eclipse TS200 epifluorescence microscope (Nikon Corporation). The corrected fluorescence intensity of the cells was measured using ImageJ software version 1.53f (National Institutes of Health) by subtracting the value of mean background fluorescence from total mean fluorescence.
Determination of the total reactive oxygen species (ROS) and mitochondrial superoxide level
The elevation of intracellular ROS induced by a cisplatin treatment was detected by DCFH-DA. Briefly, WT and MDR HCT-116 cells were seeded at 1×105 cells/well in 12-well plates and treated with 50 µM cisplatin or vehicle for 12 h at 37°C. The cells were washed twice with PBS and incubated with 10 µM of DCFH-DA for 15 min at 37°C. Fluorescent micrographs were obtained using a Nikon Eclipse TS200 fluorescence microscope (Nikon Corporation). The corrected fluorescence intensity of the cells was measured using ImageJ software version 1.53f (National Institutes of Health) by subtracting the value of mean background fluorescence from total mean fluorescence.
Accumulation of mitochondrial superoxide generation was determined using MitoSOX Red™ Mitochondrial Superoxide Indicator (Invitrogen; Thermo Fisher Scientific, Inc.). Briefly, the WT and MDR HCT-116 cisplatin- or vehicle treated cells were incubated with 5 µM of a MitoSOX indicator for 10 min at 37°C, then washed twice with PBS; images were captured using a Nikon Eclipse TS200 fluorescence microscope. Mito-ID green dye (Enzo Life Sciences, Inc.) was used to stain the mitochondria, regardless of their energetic state. The corrected fluorescence intensity of the cells was measured using ImageJ software version 1.53f (National Institutes of Health) by subtracting the value of mean background fluorescence from total mean fluorescence.
Electrophoretic mobility shifts assay (EMSA)
EMSA analyses were performed using an Electrophoretic Mobility Shift Assay kit (cat. no. E33075; Molecular Probes; Thermo Fisher Scientific, Inc.) according to the manufacturer's recommendation. Nuclear extracts (1 µg), aforementioned, were used to examine their binding capacity to the DNA-binding motif of activator protein 1 (AP1). Table SIII lists the AP1 consensus sequence used for EMSA, which was purchased from Santa Cruz Biotechnology, Inc. (AP-1 Gel Shift Oligonucleotides; cat. no. sc-2501). Briefly, 100 pmol AP1 consensus oligo was incubated with nuclear extract in 1X binding buffer (750 mM KCl; 0.5 mM dithiothreitol; 0.5 mM EDTA; 50 mM Tris, pH 7.4) at room temperature for 20 min before loading onto a 6% non-denaturing polyacrylamide gel. The EMSA bands were visualize by staining with 1X SYPRO® Ruby EMSA stain according to manufacturer protocol.
Determination of autophagy using Cyto-ID and MDC immunocytochemistry
A total of 1×105 WT and MDR cells [including Mock (empty vector), GSTO1 activation CRISPR and GSTO1 KD CRISPR transfected HCT-116 cells] were cultured on coated glass coverslip and treated with 50 µM cisplatin or vehicle for 12 h at 37°C and 5% CO2 in complete medium. Cisplatin- or vehicle-treated cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. The fixed cells were stained using the components in the CYTO-ID® Autophagy Detection Reagent (cat. no. ENZ-51031; Enzo Life Sciences, Inc.) according to the manufacturer's protocol. Cyto-ID is a proprietary reagent that labels the autophagy vacuoles specifically. DAPI was used to stain the nucleus. Fluorescence image acquisitions were performed using a Nikon Eclipse fluorescence microscope (Nikon Corporation).
MDC (MilliporeSigma; Merck KGaA) was used to stain the autophagy vacuoles in the GSTO1 KD plasmid transfected MDR cells. GSTO1 KD plasmid contains a GFP as a selection marker that restricts the use of Cyto-ID Green Detection Reagent in this case. CYTO-ID® and MDC fluorescent intensity were measured using ImageJ software version 1.53s (National Institutes of Health).
Flow cytometry assays for autophagy and apoptosis
Cell death and the development of acidic vacuoles were quantified by flow cytometry (Beckman Coulter, Inc.). WT and MDR HCT-116 cells [including Mock (empty vector), GSTO1 activation CRISPR and GSTO1 KD CRISPR transfected HCT-116 cells] were seeded at 1×105 cells/well in a 6-well plate and treated with cisplatin or vehicle for 12 h at 37°C and 5% CO2 in complete media.
The cells were harvested and washed twice with PBS. An in situ Cell Death Detection Kit (cat. no. 11684795910; Roche Applied Sciences) and CYTO-ID® Autophagy Detection Kit (Enzo Life Sciences, Inc.) were used to quantify apoptosis induction and the development of acidic vesicular organelles, respectively. Green (510-530 nm) fluorescence emission from ~5×104 cells illuminated with blue (488 nm) excitation light was measured using a Beckman Coulter Gallios flow cytometer and analyzed by Kaluza Analysis Software version 2.0 (both from Beckman Coulter, Inc.).
Immunoprecipitation and protein mass fingerprinting
The aforementioned WT and MDR HCT-116 cell lysates (1 mg in 500 µl of RIPA buffer) were immunoprecipitated with 2 µg of GSTO1 antibody, and the samples were rotated overnight at 4°C. A total of 40 µl Protein A/G Sepharose beads (Pierce™; Thermo Fisher Scientific, Inc.) were added to each sample and rotated for 1 h at 4°C. The beads were washed three times with RIPA lysis buffer, and 30 µl supernatant fractions were collected by centrifugation at 2,000 × g for 10 min and subsequently subjected to 10% SDS-PAGE followed by stained with Coomassie blue. The proteins were identified by peptide mass fingerprinting. Briefly, protein spots were excised, digested with trypsin (Promega Corporation), and mixed with α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.1% TFA, and subjected to MALDI-TOF analysis (Microflex LRF 20; Bruker Daltonics; Bruker Corporation). Spectra were collected from 300 shots/spectrum over the m/z range 600-3000 and calibrated by two-point internal calibration using Trypsin auto-digestion peaks (m/z 842.5099, 2211.1046). The peak list was generated using Flex Analysis 3.0 (Bruker Daltonics; Bruker Corporation). The threshold used for peak-picking was as follows: 500 for a minimum resolution of monoisotopic mass, 5 for the signal to background noise. The MASCOT online database (Matrix Science, Ltd.) was used for protein identification. The following parameters were used for the database search: Trypsin as the cleaving enzyme; a maximum of one missed cleavage; iodoacetamide (Cys) as a complete modification; oxidation (Met) as a partial modification; monoisotopic masses; and a mass tolerance of ±0.1 Da.
Generation of GSTO1-activated, GSTO1-knockdown (KD) and TNFαIP3/A20-KD cells using CRISPR-Cas9
CRISPR-Cas9-mediated genome editing was performed using the GSTO1 CRISPR-Cas9 KD human plasmid (cat. no. sc-404107), TNFαIP3/A20 CRISPR-Cas9 KD human plasmid (cat. no. sc-400447-KO-2) and GSTO1 CRISPR activation human plasmid (cat. no. sc-404107-ACT) purchased from Santa Cruz Biotechnology, Inc. Human GSTO1 KD CRISPR plasmid mixture is a pool of 3 different guide (g)RNA plasmids, each encoding the Cas9 nuclease and a target-specific 20 nucleotide (nt) gRNA in the GSTO1 gene: gRNA 1 (5′-GCG TCT AGT CCT GAA GGC CA-3′), which targets exon 2 and N-terminal domain of the protein; gRNA 2 (5′-ACA ACT CTA AGA TCA TCT TC-3′), which targets exon 3 and N-terminal domain of the protein; and gRNA 3 (5′-TCTAATAAAGCTTCCTACCA-3′), which targets exon 6 and C-terminal domain of the protein GSTO1. CRISPR/Cas9 KD plasmid disrupted gene expression by causing a double-strand break in a 5′ constitutive exon within the GSTO1 gene. A single gRNA (5′-AGG TCA GTG TCA CGG GAG GG-3′) sequence was used for human GSTO1 CRISPR activation plasmid. The human GSTO1 CRISPR activation plasmid is a synergistic activation-mediator (SAM) transcription activation system that specifically upregulates the GSTO1 gene. The GSTO1 CRISPR activation plasmid solution contains an equimolar ratio of the following three plasmids: CRISPR/dCas9-VP64-Blast plasmid, which encodes the deactivated Cas9 (dCas9) nuclease fused to the transactivation domain VP64 and a blasticidin resistance gene; MS2-P65-HSF1-Hygro plasmid, which encodes the MS2-p65-HSF1 fusion protein and a hygromycin resistance gene; sgRNA (MS2)-Puro plasmid, which encodes a target-specific 20 nt gRNA and a puromycin resistance gene. The SAM complex activates transcription of GSTO1 and upregulates GSTO1 gene expression.
Human TNFαIP3/A20 KD CRISPR plasmid is also a pool of 3 different gRNA plasmids: gRNA 1 (5′-CAC GCA ACT TTA AAT TCC GC-3′), which targets exon 3 and TRAF-binding domain of the protein; gRNA 2 (5′-GTG AAC GTT GCC ACA ACG CC-3′), which targets exon 7 and TNIP1-binding domain of the protein; and gRNA 3 (5′-CTT GTG GCG CTG AAA ACG AA-3′), which targets exon 2 and TRAF-binding domain of the protein. TNFαIP3/A20 CRISPR/Cas9 KD plasmid disrupted gene expression by causing a double-strand break in a 5′ constitutive exon within the TNFαIP3/A20 gene. A total of 1×106 WT and MDR HCT-116 cells were seeded on a 6-well plate in antibiotic free complete media. The cells were transfected with 1 µg of CRISPR plasmid mixture using UltraCruz® transfection reagent (Santa Cruz Biotechnology, Inc.) for 24 h at 37°C. Transfection media was replaced with complete growth media and cells were incubated at 37°C for an additional 48 h before selection using 1 µg/ml puromycin (MilliporeSigma; Merck KGaA). Cells were selected for 2-4 weeks, dependent on growth recovery. Western blotting was used to confirm the KD and activation efficiency. An empty vector was used as the Mock group in both GSTO1-activation and -KD experiments.
Statistical analysis
All data are presented as means ± SD using an unpaired Student's t-test or one-way ANOVA (with Tukey's post hoc test for multiple comparisons) using GraphPad Prism 9.0 software (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference. Each experiment was performed at least three times unless indicated otherwise. Each western blot was performed in duplicate.
Results
MDR HCT-116 cells exhibit higher GSTO1 expression and reduced cisplatin sensitization
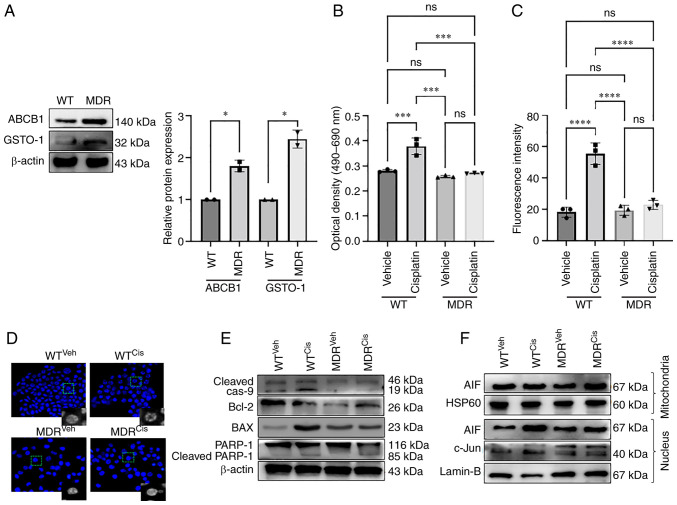
ABCB1 expression is increased in cancer cells during the development of anti-cancer drug resistance, which facilitates the efflux of drugs from the cell (17). ABCB1 protein expression was increased in MDR HCT-116 cells compared with the WT cells as determined by western blotting (Fig. 1A); GSTO1 expression was also increased in the MDR cells compared with WT cells. MDR HCT-116 cells showed resistance to the 50 µM cisplatin treatment compared with WT cells, as determined by an LDH assay (Fig. 1B) and apoptosis assay using flow-cytometry (Figs. 1C, S1E, S1F and S2). Cisplatin treatment significantly increased LDH release in WT cells compared with the vehicle-treated cells (Fig. 1B). LDH release in cisplatin-treated MDR cells was also significantly higher compared with vehicle-treated MDR cells, although the effectiveness of cisplatin was much lower in MDR cells compared with the WT cells (Fig. 1B). Moreover, cisplatin treatment increased apoptosis significantly in WT cells compared with the vehicle-treated cells, but the effect was lower in MDR cells (Figs. 1C, S1E, S1F and S2).
Figure 1.
MDR HCT-116 cells are resistant to cisplatin-induced apoptosis. (A) The protein expression levels of the ABCB1 and GSTO1 proteins in WT and MDR HCT-116 cells were analyzed by western blotting and quantified using ImageJ software. Unpaired Student's t-test was used for statistical analysis. WT and MDR cancer cells were treated with 50 µM cisplatin for 12 h, and cell apoptosis was analyzed by (B) LDH assay and (C) ApoStrand™ apoptosis assay; one-way ANOVA was used for statistical analysis. (D) Nuclear damage was analyzed by Hoechst 33342 staining (×200 magnification). (E) Western blotting of apoptosis markers. (F) Mitochondrial and nuclear proteins were analyzed by western blotting to determine the expression of AIF and c-Jun proteins. *P<0.05; ***P<0.001; ****P<0.0001. AIF, apoptosis-inducing factor; cas, caspase; cis, cisplatin; GSTO1, glutathione-S-transferase Ω-1; HSP, heat-shock protein; LDH, lactate dehydrogenase; MDR, multidrug resistant; MDR1, multidrug-resistance protein 1; ns, not significant; PARP, poly(ADP-ribose) polymerase 1; veh, vehicle; WT, wild-type.
Cisplatin-treated WT cells showed morphological deformities, mostly membrane blebbing compared with the vehicle-treated WT, MDR and cisplatin-treated MDR cells (Fig. S1A). Total ROS generation was increased in both cisplatin-treated WT and MDR cells compared with their respective vehicle-treated cells; however, no significant difference was noted in the cisplatin-treated MDR cells compared with the cisplatin-treated WT cells (Fig. S1B). The mechanism of cisplatin-mediated cytotoxicity is dependent on DNA-adduct formation; that is, DNA damage-mediated cell death (18). Fig. 1D shows severe chromatin decondensation in the cisplatin-treated WT cells, which was not visible in the MDR or cisplatin-treated MDR cells. DNA damage by cisplatin treatment leads to mitochondria-mediated apoptosis (18-20). The total ROS generation following cisplatin treatment led to an increase in ROS in the mitochondria of WT and MDR cells compared with their respective vehicle-treated controls; however, no significant difference was noted between the two cisplatin-treated groups (Fig. S1C and D).
Cisplatin treatment increased the cleavage of caspase 9 and poly(ADP-ribose) polymerase 1, and increased Bax expression in the WT cells compared with the vehicle-treated WT cells and MDR cells (Figs. 1E and S3A). Expression of cleaved PARP-1 was also significantly higher in cisplatin-treated MDR cells compared with the vehicle-treated MDR cells, although the level was much lower than that of cisplatin-treated WT cells (Figs. 1E and S3A). Bcl2 expression levels was not significantly different between vehicle-treated WT cells and cisplatin-treated WT cells, nor between cisplatin-treated WT cells and cisplatin-treated MDR cells (Figs. 1E and S3A). Moreover, Bcl2 expression was significantly reduced in vehicle and cisplatin-treated MDR cells compared with the vehicle-treated cells (Figs. 1E and S3A). A previous publication also demonstrated a reduction of Bcl2 expression during autophagy activation (21). The translocation of the apoptosis-inducing factor (AIF) from the mitochondria to the nucleus induces apoptosis in drug-treated cancer cells (22). The nuclear/mitochondrial expression ratio of AIF was significantly increased in the cisplatin-treated WT cells compared with the vehicle-treated WT cells and cisplatin-treated MDR cells (Figs. 1F, S3B and C). Cisplatin treatment also reduced c-Jun nuclear expression (Figs. 1F and S3D) and binding of AP1 transcription factor with its consensus DNA sequence in the cisplatin-treated MDR cells compared with the cisplatin-treated WT cells (Fig. S4A and B), a well-documented mode of action of cisplatin-mediated cell death (23). AP1 binding with its DNA sequence was also reduced in vehicle-treated compared with the cisplatin-treated WT cells. Moreover, cisplatin-treated WT cells showed similar AP1 binding as seen in vehicle and cisplatin-treated MDR cells (Figs. S4A and B).
MDR HCT-116 cells show an unfolded protein response (UPR)-mediated autophagy
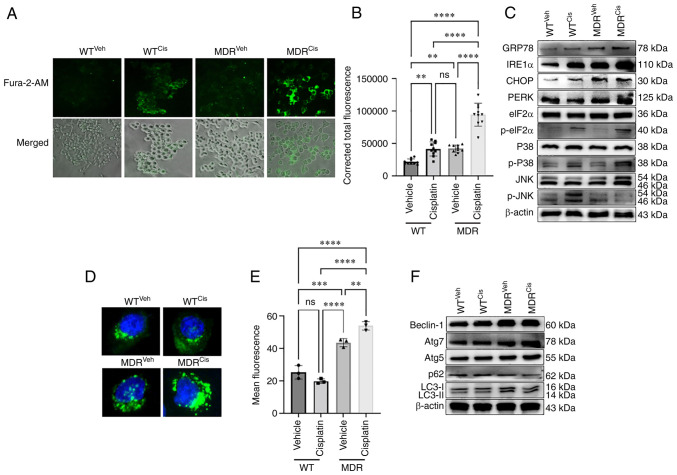
Previous studies suggest that the elevated levels of mitochondrial ROS can promote both autophagy and apoptosis (9,24,25). Cisplatin-treatment activated calcium signaling in MDR cells (Fig. 2A and B). Significantly higher calcium deposition was noted in the cisplatin-treated MDR cells compared with the vehicle-treated MDR and the cisplatin-treated WT cells (Fig. 2A and B). Cellular calcium level was also higher in cisplatin-treated WT cells compared with the vehicle-treated WT cells (Fig. 2A and B), although that level was much lower than in cisplatin-treated MDR cells. A previous study demonstrated that cellular calcium level determined cellular fate towards apoptosis or autophagy (26). High calcium can activate ROS in mitochondria (27), similar to what was observed in cisplatin-treated WT cells (Fig. 2A and B). Calcium-induced UPR is a well-studied mechanism that ultimately leads to autophagy (24,28). Expression levels of the UPR protein markers glucose-regulated protein 78 (GRP78), CHOP and PKR-like endoplasmic reticulum kinase (PERK) were increased in the cisplatin-induced MDR cells compared with the cisplatin-induced WT cells (Figs. 2C and S5A). Interestingly, no significant difference was noted when inositol-requiring kinase 1α (IRE1α) expression was compared between cisplatin-treated MDR and WT cells (Figs. 2C and S5A). The present study also noticed a significantly higher expression of IRE1α and CHOP in cisplatin-treated WT cells compared with the vehicle-treated WT cells, suggesting a mild ER stress mediated-apoptosis (29,30). No significant difference in GRP78, IRE1α, CHOP and PERK protein expression levels were noted in vehicle-compared with cisplatin-treated MDR cells (Figs. 2C and S5A).
Figure 2.
MDR HCT-116 cells exhibit an increase in calcium signaling-dependent UPR and autophagy. WT and MDR HCT-116 cells were treated with 50 µM cisplatin or vehicle for 12 h and subsequently (A) stained with Fura-2/AM to determine the cellular calcium release (×200 magnification) and (B) quantified using ImageJ (n=400 cells); one-way ANOVA was used for statistical analysis. (C) Western blotting analysis of ER stress- and UPR-related markers in WT and MDR cells with or without cisplatin exposure. (D) Fluorescence images of autophagic vesicles stained with Cyto-ID® Autophagy reagent (green) in WT and MDR cells (×1,000 magnification); nuclei were stained with Hoechst 33342 (blue). (E) Quantification of fluorescence intensity presented of cells presented in part (D); one-way ANOVA was used for statistical analysis. (F) Representative immunoblots of autophagy marker protein expressions in whole-cell protein lysates. **P<0.01; ***P<0.001; ****P<0.0001. ATG, autophagy-related gene; cis, cisplatin; eIF2α, eukaryotic initiation factor 2α; ER, endoplasmic reticulum; GRP78, glucose-regulated protein 78; IRE1α, inositol-requiring kinase 1α; MDR, multidrug resistant; ns, not significant; p-, phosphorylated; PERK, PKR-like endoplasmic reticulum kinase; UPR, unfolded protein response; veh, vehicle; WT, wild-type.
The phosphorylation of p38 and eukaryotic initiation factor 2α (eIF2α) were reported as positive markers for ER stress-mediated UPR mechanism, even though both are also expressed during apoptosis (31,32). p38 and eIF2α proteins can be phosphorylated during activation of apoptosis or autophagy (33,34). The phosphorylation of eIF2α was higher in the cisplatin-treated WT and MDR cells compared with the vehicle-treated WT and MDR cells (Figs. 2C, S5A and B). Interestingly, phosphorylation of p38 showed no significant difference between vehicle and cisplatin-treated WT cells nor between cisplatin-treated WT and MDR cells. However, p-p38/p38 ratio was significantly higher in cisplatin-treated MDR cells compared to the cisplatin-treated WT cells (Figs. 2C, S5A and B). JNK phosphorylation was lower in the cisplatin-treated MDR cells compared with the cisplatin-treated WT cells (Figs. 2C, S5A and B).
Furthermore, immunofluorescence and flow cytometry confirmed an increase in autophagic flux in MDR cells compared with WT cells, regardless of the treatments. A significant increase in autophagy activation was observed in vehicle- and cisplatin-treated MDR cells compared with the vehicle- and cisplatin-treated WT cells (Figs. 2D and E); autophagic flux was higher upon cisplatin treatment compared with vehicle treatment in MDR cells. Similarly, flow cytometry also demonstrated higher autophagic flux in MDR cells compared with the WT cells irrespective of treatment. Notably, cisplatin treatment further increased autophagic flux in MDR cells compared with in vehicle-treated MDR cells (Figs. S6A, S6B and S7). The protein expression levels of autophagy markers Beclin-1, autophagy-related gene (ATG)-7, ATG5 and LC3II increased, whereas the expression of ubiquitin-binding protein p62 (p62; also known as sequestosome-1) decreased in cisplatin-treated MDR cells compared with the cisplatin-treated WT cells (Figs. 2F, S6C and D). Treatment with autophagy inhibitor BAF-A1 sensitized the MDR cells to cisplatin, inducing the expression of apoptosis and depleting the levels of autophagy markers (Fig. S8). BAF-A1 treatment significantly increased p62 expression in MDR cells compared with the vehicle-only- and cisplatin-only-treated group (Fig. S8A and B); however, BAF-A1 and cisplatin co-treatment further increased p62 level compared with all other groups. LC3II/LC3I ratio was decreased in BAF-A1-treated cells with or without cisplatin co-treatment compared with the vehicle-only- and cisplatin-only-treated cells (Figs. S8A and C). Moreover, BAF-A1 and cisplatin co-treatment increased expression levels of the apoptosis markers cleaved caspase 9, cleaved caspase 3 and cytochrome c compared with the vehicle, only BAF-A1and only cisplatin-treated cells (Figs. S8A and D-F).
CRISPR-Cas9 activation of GSTO1 in WT HCT-116 cells activates autophagy
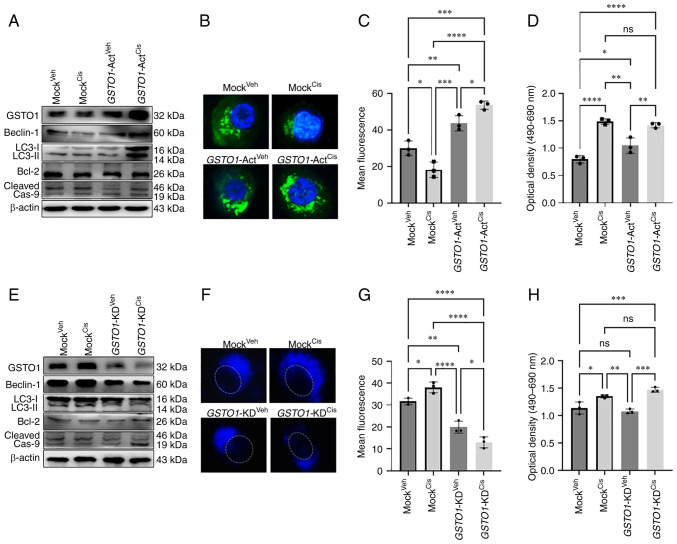
To mimic the drug-resistance condition in WT cells, GSTO1 was activated and checked to determine if it could induce autophagy. Figs. 3A and S9E demonstrated that CRISPR-Cas9 genome editing successfully induced GSTO1 expression in vehicle- and cisplatin-treated WT cells compared with the Mock groups. GSTO1 activation induced the expression of autophagy marker proteins Beclin-1 and LC3II in cisplatin-treated, GSTO1-activated WT cells compare with the vehicle- or cisplatin-treated Mock cells (Figs. 3A and S10A). Interestingly, Beclin-1 and LC3II expression levels were significantly higher in cisplatin-treated, GSTO1-activated cells compared with the vehicle-treated, GSTO1-activated cells, which could be attributed to a connection between GST-mediated drug-efflux and autophagy activation (35). Apoptosis marker cleaved caspase 9 was significantly reduced marginally in the GSTO1-activated, cisplatin-treated WT cells compared with the cisplatin-treated Mock WT cells (Figs. 3A and S10A).
Figure 3.
GSTO1 activation in WT HCT-116 cells induces autophagy following cisplatin treatment, whereas GSTO1 inhibition in MDR cells induces apoptosis. GSTO1 was overexpressed using GSTO1-Act CRISPR plasmid transfected in WT HCT-116 cells. (A) Western blotting was performed to determine the protein expression levels of GSTO1 expression as well as autophagy and apoptosis marker proteins. (B) Fluorescence images of autophagic vesicles stained with Cyto-ID® Autophagy reagent (green) in Mock- and GSTO1-Act plasmid-transfected WT cells (×1,000 magnification); nuclei are stained with DAPI (blue). (C) Quantification of fluorescence intensity of the data presented in part (B); one-way ANOVA was used for statistical analysis. (D) Cell death was analyzed using an LDH ELISA assay; one-way ANOVA was used for statistical analysis. GSTO1 was inhibited using the GSTO1-KD CRISPR/Cas9 plasmid in MDR cells. (E) Western blotting was used to assess protein expression levels of GSTO1 as well as autophagy and apoptosis marker proteins. (F) Representative fluorescence images of autophagic vesicles stained with monodansylcadaverine (blue); the dashed white line indicates the outline of the nucleus (×1,000 magnification). (G) Quantification of fluorescence intensity of the data presented part (F); one-way ANOVA was used for statistical analysis. (H) Cell death was analyzed using an LDH ELISA assay; one-way ANOVA was used for statistical analysis. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. Act, activation; cas, caspase; cis, cisplatin; GSTO1, glutathione-S-transferase Ω-1; KD, knockdown; LDH, lactate dehydrogenase; MDR, multidrug resistant; ns, not significant; veh, vehicle; WT, wild-type.
Unexpectedly, Beclin-1 expression was reduced considerably in cisplatin-treated Mock CRISPR-transfected cells (Figs. 3A and S10A) compared with WT cisplatin-treated cells (Figs. 2F and S6C). Notably, a slight increase in the number of floating cells in the Mock cisplatin-treated CRISPR-transfected group compared with the WT cisplatin-treated group (data not shown). Presumably, the puromycin selection procedure might increase cisplatin sensitivity in Mock CRISPR cells compared with WT cells, leading to necroptosis and decreased Beclin-1 expression. However, a recent study also revealed that puromycin selection might occasionally impede protein expression (36).
Immunofluorescence and flow cytometric analysis showed that GSTO1 activation and cisplatin treatment resulted in higher autophagy flux compared with that observed in the cisplatin-treated Mock cells. Cisplatin treatment produced more autophagic vesicles in GSTO1-activated cells compared with the cisplatin-treated Mock cells (Figs. 3B, 3C, S9C, S9D and S11). The LDH assay revealed a slight reduction of apoptosis in the cisplatin-treated, GSTO1-activated cells compared with the cisplatin-treated Mock cells, but the difference was not significant (Fig. 3D). Cisplatin treatment induced significant amount of apoptotic cell death in Mock group compared with the vehicle-treated Mock and GSTO1-ACT groups (Fig. 3D). Protein expression levels of the UPR-related protein markers GRP78, IRE1α and PERK were not changed in either cisplatinor vehicle-treated GSTO1 activated cells compared with the cisplatin- or vehicle-treated Mock cells (Fig. S9A and B).
CRISPR-Cas9 inhibition of GSTO1 in MDR HCT-116 cells inhibits autophagy and sensitizes them to cisplatin treatment
Figs. 3E and S9E confirmed the GSTO1 inhibition efficiency in MDR cells. Although GFP fluorescence showed a satisfactory transfection efficiency of Control and GSTO1-KD CRISPR-Cas9 plasmid (Fig. S9F), residual GSTO1 was detected in western blotting analysis. Therefore, we used the term 'knockdown', rather than knockout. The inhibition of GSTO1 in MDR cells reduced the expression levels of the autophagy markers Beclin-1 and LC3II compared with their expressions in the Mock MDR cells (Figs. 3E and S10B). Protein expression of the apoptosis marker cleaved caspase 9 was increased in GSTO1-inhibited cisplatin-treated MDR cells compared with the cisplatin-treated Mock MDR cells (Figs. 3E and S10B). Consistent with the western blotting results, autophagy was also reduced in the GSTO1-KD MDR cells compared with the Mock MDR group (Fig. 3F and G). The LDH assay further confirmed the sensitization of GSTO1-inhibited MDR cells to cisplatin (Fig. 3H). Cisplatin-treatment significantly induced LDH release in GSTO1-KD cells compared with the cisplatin-treated Mock cells, a vehicle-treated Mock or GSTO1-KD cells (Fig. 3H). GSTO-KD and cisplatin treatment in MDR cells inhibited GRP78, IRE1α and PERK compared with the Mock cisplatin-treated cells (Fig. S9G and H).
GSTO1-TNFαIP3/A20 interaction is a novel mechanism towards drug resistance
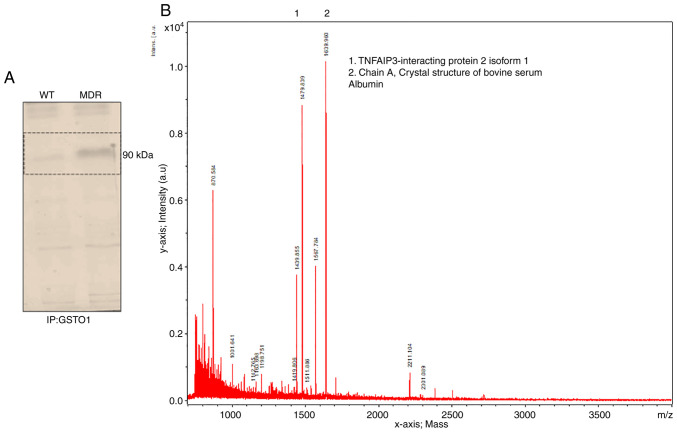
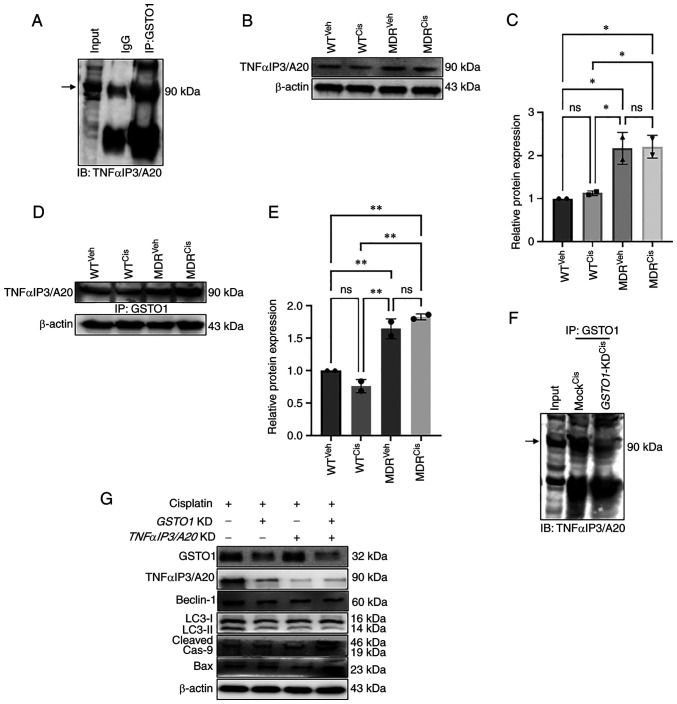
The GSTO1 pull-down assay showed an interacting protein band in MDR cells but not in WT cells (Fig. 4A). Subsequently, MALDI-TOF mass spectrometry identified TNFαIP3/A20 as a putative novel binding partner of GSTO1 in MDR cells (Fig. 4B). Immunoprecipitation of the GSTO1 protein and subsequent immunoblotting for TNFαIP3/A20 in MDR cells confirmed an interaction between GSTO1 and TNFαIP3/A20 (Fig. 5A). TNFαIP3/A20 expression was increased in both vehicle- and cisplatin-treated MDR cells compared with the vehicle- and cisplatin-treated WT cells (Fig. 5B and C). Moreover, interaction with GSTO1 also increased in the MDR cells compared with the WT cells, particularly following cisplatin treatment (Fig. 5D and E). Furthermore, TNFαIP3/A20 and GSTO1 interaction was reduced in GSTO1-KD MDR cells compared with Mock-transfected cells (Fig. 5F). Western blotting analysis showed that TNFαIP3/A20 protein expression may be dependent on the GSTO1 expression status, as significantly reduced TNFαIP3/A20 expression was detected in the GSTO1-KD cells (Figs. 5G and S12). GSTO1 and TNFαIP3/A20 single-gene or double-gene KD combined with cisplatin co-treatment showed significantly reduced Beclin-1 expression compared with cisplatin-treated Mock cells. No significant differences were identified between GSTO1 and TNFαIP3/A20 single-gene KD and double-gene KD groups (Figs. 5G and S12). LC3II/LC3I ratio was also reduced significantly in GSTO1 and TNFαIP3/A20 single-gene or double-gene KD cisplatin-treated cells compared with cisplatin-treated Mock cells (Figs. 5G and S12). GSTO1 and TNFαIP3/A20 double-gene KD cisplatin-treated cells showed significantly lower LC3II/LC3I ratio compared with GSTO1 KD cisplatin-treated cells. Expression levels of cleaved caspase 9 were increased in GSTO1 single-gene KD or GSTO1 and TNFαIP3/A20 double-KD cisplatin-treated cells compared with the cisplatin-treated Mock cells (Figs. 5G and S12); no significant difference was identified between the cisplatin-treated TNFαIP3/A20-KD and the cisplatin-treated Mock groups. GSTO1 and TNFαIP3/A20 double-gene KD cisplatin-treated cells showed significantly higher cleaved caspase 9 expression compared with the GSTO1 and TNFαIP3/A20 single-gene KD cisplatin-treated cells (Figs. 5G and S12). Notably, only GSTO1 and TNFαIP3/A20 double-gene KD cisplatin-treated cells showed significantly higher Bax expression compared with the other three groups (Figs. 5G and S12). Overall, the results suggested that the GSTO1-TNFαIP3/A20 axis may be important for drug resistance in colon cancer cells, and the inhibition of this axis may sensitize MDR cells to cisplatin.
Figure 4.
GSTO1 interacts with a novel partner, TNFαIP3/A20, in MDR HCT-116 cells. (A) GSTO1 pull-down from whole cell lysates were subjected to SDS-PAGE and stained with Coomassie Blue. The protein band was excised and determined by MALDI-TOF. (B) MALDI-TOF analysis determined TNFαIP3/A20 and bovine serum albumin (chain A) as GSTO1 interacting partners. A.u., arbitrary units; GSTO1, glutathione-S-transferase Ω-1; MALDI-TOF, matrix-assisted laser desorption/ionization-time of flight; MDR, multidrug resistant; ns, not significant; TNFαIP3/A20, TNF-α-induced protein 3/zinc-finger protein A20; WT, wild-type.
Figure 5.
Inhibition of GSTO1-TNFαIP3/A20 interaction sensitizes MDR HCT-116 cells to cisplatin. (A) Co-IP was used to determine the interaction of endogenous GSTO1 and TNFαIP3/A20 in the whole cell lysate of MDR HCT116 cells. (B) Western blotting and (C) densitometric analysis showing that TNFαIP3/A20 expression was increased in MDR cells compared with the WT cells irrespective of treatments; one-way ANOVA was used for statistical analysis. (D) GSTO1 pull-down, western blotting for TNFαIP3/A20 and (E) densitometric analysis showing that the GSTO1-TNFαIP3/A20 interaction is increased in MDR cells compared with WT cells irrespective of treatments; one-way ANOVA was used for statistical analysis. (F) GSTO1 co-IP in cis-treated Mock and GSTO1-KD MDR cells shows that the GSTO1-TNFαIP3/A20 interaction is reduced in GSTO1-KD cells. (G) Representative western blotting images of GSTO1, TNFαIP3/A20 and apoptosis- and autophagy-related protein markers in GSTO1 and TNFαIP3/A20 single-KD and double-KD MDR cells showing that the double KD had a more notable effect compared with either single KD. *P<0.05; **P<0.01. GSTO1, glutathione-S-transferase Ω-1; cas, caspase; cis, cisplatin; IB, immunoblot; IP, immunoprecipitation; KD, knockdown; MDR, multidrug resistant; ns, not significant; TNFαIP3/A20, TNF-α-induced protein 3/zinc-finger protein A20; veh, vehicle; WT, wild-type.
Discussion
Cisplatin is commonly used as a chemotherapeutic agent in a number of cancers, including colon cancer (37). Cisplatin induces oxidative damage of DNA, mitochondrial apoptosis, and cell death (38). However, some cancers, including colon, lung and ovarian cancer, can acquire resistance to cisplatin chemotherapy, leading to treatment failure (39-41). Cancer cell resistance to a particular drug is often accompanied by resistance to other drugs, a condition known as multidrug resistance, which significantly reduces cancer chemotherapy efficacy and overall survival of patients (42). GST family enzymes are responsible for acquired resistance either by drug efflux or drug inactivation; GST family proteins protect cellular macromolecules from oxidants (43,44). A previous study showed that GSTs inactivate cisplatin by thiol conjugation (18). Although isoforms of GST enzyme have been reported to serve a role in drug resistance (5-7), little is known about the role of GSTO1 in drug resistance. Also, there are no previous reports showing that GSTO1 is increased in chemoresistant colon cancer tissues. In this study, the molecular mechanism of GSTO1-mediated drug resistance was investigated. The results demonstrated a higher expression of both ABCB1 and GSTO1 in MDR cells compared with WT HCT-116 cells. Although cisplatin effectively induced the total cellular ROS and mitochondrial ROS in both WT and MDR cells, the ROS in WT cells led to mitochondria-dependent apoptosis, whereas it activated calcium signaling in MDR. Interestingly, transcription factor AP1 showed significantly higher binding with its DNA sequence in cisplatin-treated WT, MDR and vehicle-treated MDR cells compared with the vehicle treated WT cells. AP1 is a transcription factor that binds with consensus DNA sequences across the genome and AP1 binding with its consensus DNA increased upon apoptosis activation (19). A previous publication also documented that AP1 is essential for autophagosomes formation (45).
We hypothesized that calcium signaling may aid MDR cells bypass cisplatin by activating UPR-mediated autophagy activation. Therefore, the role of calcium in chemoresistant colon cancer needs to be studied in further detail, including GSTO1 knockout in an animal model.
The overexpression of GSTO1 in WT cells induced autophagy-mediated cell survival following cisplatin treatment. Therefore, targeting GSTO1 in drug-resistant cells could be an attractive target for resensitizing MDR colon cancer cells to cisplatin. A recent publication demonstrated that YTH N6-methyladenosine RNA binding protein 1 resensitized drug-resistant colon cancer cells to cisplatin (46). A recent report documented a novel role of GSTP1 in adriamycin resistance in breast cancer cells by activating autophagy (47). The overexpression of GSTP1 in MCF-7 cells activated autophagy, whereas inhibition of GSTP1 reduced autophagy in drug-resistant cells (47). Furthermore, GSTO1 inhibition in MDR cells inhibits autophagy through UPR modulation and sensitizes the cells to cisplatin. Hence, GSTO1 may be an important target for reversing drug resistance in MDR cells.
The present study results indicated that, in MDR HCT-116 cells, GSTO1 interacts with TNFαIP3/A20, and that TNFαIP3/A20 was reduced in GSTO-KD MDR cells. GSTO1 expression may regulate TNFαIP3/A20 expression in MDR cells. However, GSTO1 expression was not dependent on the TNFαIP3/A20 status in cells. GSTO1-TNFαIP3/A20 double-KD MDR cells were more effective than either GSTO1 or TNFαIP3/A20 single KD in lowering the expression levels of apoptosis markers upon cisplatin treatment. TNFαIP3/A20 is a ubiquitin-editing enzyme that negatively regulates NF-κB and plays a major role in the biology of cells (48,49). Notably, a previous study suggested that TNFαIP3/A20 mediates the resistance to DNA-damage repair in cancer (50). Furthermore, a previous publication also suggested that TNFαIP3/A20 is a major factor in tamoxifen resistance in breast cancer cells (51).
The contribution of the GST family proteins to cancer and chemoresistance is well known, but the available information on GSTO1 is unclear. The present study examined a novel interaction between GSTO1 and TNFαIP3/A20. The results suggested that targeting both is more effective than targeting either alone, in relation to chemosensitivity. Including specific inhibitors for both GSTO1 and TNFαIP3/A20 in chemotherapy may help prevent drug resistance, but a detailed mechanistic study on mice or human subjects is required. In future studies, GSTO1 and TNFαIP3/A20 double-knockout mice will be generated to examine acquired drug resistance in an in vivo colon cancer model. Furthermore, a detailed study on the role of the inflammatory pathway related to TNFαIP3/A20 in chemoresistant cancer cells is also required because TNFαIP3/A20 is an inflammation pathway-related protein (49).
In conclusion, the GSTO1-TNFαIP3/A20 interaction may be vital during acquired drug resistance. A previous study reported TNFαIP3/A20 promotes survival of CD4 T cells by promoting autophagy (52); TNFαIP3/A20 was also overexpressed in HCT-116 MDR cells in the present study. Both GSTO1 and TNFαIP3/A20 were reported to be upregulated during drug resistance (10-12,50,51). The present study identified a novel interaction between these two proteins in acquired MDR cells. Therefore, targeting the GSTO1-TNFαIP3/A20 axis may help overcome MDR. 4. S13 represents a schematic diagram of the mechanism of GSTO1-mediated drug resistance in colon cancer cells. WT HCT-116 cells showed apoptotic cell death upon cisplatin treatment, whereas MDR cells survived upon cisplatin treatment by activating autophagy. GSTO1 was upregulated and interacted with TNFαIP3/A20 in MDR cells. Activation of GSTO1 in WT cells activated autophagy and inhibition of GSTO1 in MDR cells sensitized them to cisplatin treatment and leading to apoptosis.
Supplementary Data
Acknowledgments
Not applicable.
Abbreviations
- ATG
autophagy-related gene
- AP1
activator protein 1
- eIF2α
eukaryotic initiation factor 2α
- ER
endoplasmic reticulum
- GRP78
glucose-regulated protein 78
- GSTO1
glutathione-S-transferase Ω-1
- IRE1α
inositol-requiring kinase 1α
- MDR
multi-drug resistant
- p62
ubiquitin-binding protein p62
- PERK
PKR-like endoplasmic reticulum kinase
- TNFαIP3/A20
TNF-α-induced protein 3/zinc-finger protein A20
- UPR
unfolded protein response
Funding Statement
This study was partially supported by the Daegu University Research Grant, 2020.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SP designed the study, performed experiments, interpreted results and wrote the manuscript. MB contributed to performing western blotting and data analysis. SCK designed the study, interpreted the results and reviewed and edited the manuscript. All authors have read and approved the final manuscript. SP, MB and SCK confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wang X, Zhang H, Chen X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019;2:141–160. doi: 10.20517/cdr.2019.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukowski K, Kciuk M, Kontek R. Mechanisms of multidrug resistance in cancer chemotherapy1. Int J Mol Sci. 2020;21:3233. doi: 10.3390/ijms21093233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köberle B, Schoch S. Platinum complexes in colorectal cancer and other solid tumors. Cancers (Basel) 2021;13:2073. doi: 10.3390/cancers13092073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mele L, Del Vecchio V, Liccardo D, Prisco C, Schwerdtfeger M, Robinson N, Desiderio V, Tirino V, Papaccio G, La Noce M. The role of autophagy in resistance to targeted therapies. Cancer Treat Rev. 2020;88:102043. doi: 10.1016/j.ctrv.2020.102043. [DOI] [PubMed] [Google Scholar]
- 5.Dang DT, Chen F, Kohli M, Rago C, Cummins JM, Dang LH. Glutathione S-transferase pi1 promotes tumorigenicity in HCT116 human colon cancer cells. Cancer Res. 2005;65:9485–9494. doi: 10.1158/0008-5472.CAN-05-1930. [DOI] [PubMed] [Google Scholar]
- 6.Ogino S, Konishi H, Ichikawa D, Matsubara D, Shoda K, Arita T, Kosuga T, Komatsu S, Shiozaki A, Okamoto K, et al. Glutathione S-transferase Pi 1 is a valuable predictor for cancer drug resistance in esophageal squamous cell carcinoma. Cancer Sci. 2019;110:795–804. doi: 10.1111/cas.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su F, Hu X, Jia W, Gong C, Song E, Hamar P. Glutathion S transferase pi indicates chemotherapy resistance in breast cancer. J Surg Res. 2003;113:102–108. doi: 10.1016/S0022-4804(03)00200-2. [DOI] [PubMed] [Google Scholar]
- 8.Board PG, Coggan M, Chelvanayagam G, Easteal S, Jermiin LS, Schulte GK, Danley DE, Hoth LR, Griffor MC, Kamath AV, et al. Identification, characterization, and crystal structure of the omega class glutathione transferases. J Biol Chem. 2000;275:24798–24806. doi: 10.1074/jbc.M001706200. [DOI] [PubMed] [Google Scholar]
- 9.Paul S, Jakhar R, Bha rdwaj M, Kang SC. Glutathione-S-transferase omega 1 (GSTO11) acts as mediator of signaling pathways involved in aflatoxin B1-induced apoptosis-autophagy crosstalk in macrophages. Free Radic Biol Med. 2015;89:1218–1230. doi: 10.1016/j.freeradbiomed.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Lu H, Chen I, Shimoda LA, Park Y, Zhang C, Tran L, Zhang H, Semenza GL. Erratum: Chemotherapy-induced Ca2+ release stimulates breast cancer stem cell enrichment. Cell Rep. 2017;18:1946–1957. doi: 10.1016/j.celrep.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Yan XD, Pan LY, Yuan Y, Lang JH, Mao N. Identification of platinum-resistance associated proteins through proteomic analysis of human ovarian cancer cells and their platinum-resistant sublines. J Proteome Res. 2007;6:772–780. doi: 10.1021/pr060402r. [DOI] [PubMed] [Google Scholar]
- 12.De Boussac H, Kassambara A, Machura A, Chemlal D, Gourzones C, Requirand G, Robert N, Vincent L, Herbaux C, Bruyer A, Moreaux J. Genomic characterization of in vitro acquired-resistance to proteasome inhibitors. Blood. 2021;138(Suppl 1):S2651. doi: 10.1182/blood-2021-148584. [DOI] [Google Scholar]
- 13.Mowers EE, Sharifi MN, Macleod KF. Autophagy in cancer metastasis. Oncogene. 2017;36:1619–1630. doi: 10.1038/onc.2016.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao W, Wei W, Zhan Z, Xie D, Xie Y, Xiao Q. Regulation of drug resistance and metastasis of gastric cancer cells via the microRNA647-ANK2 axis. Int J Mol Med. 2018;41:1958–1966. doi: 10.3892/ijmm.2018.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget. 2014;5:8027–8038. doi: 10.18632/oncotarget.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhardwaj M, Cho HJ, Paul S, Jakhar R, Khan I, Lee SJ, Kim BY, Krishnan M, Khaket TP, Lee HG, Kang SC. Vitexin induces apoptosis by suppressing autophagy in multi-drug resistant colorectal cancer cells. Oncotarget. 2017;9:3278–3291. doi: 10.18632/oncotarget.22890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Seebacher N, Shi H, Kan Q, Duan Z. Novel strategies to prevent the development of multidrug resistance (MDR) in cancer. Oncotarget. 2017;8:84559–84571. doi: 10.18632/oncotarget.19187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Novel strategies to prevent the development of multidrug resistance (MDR) in cancer. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 19.Siddik ZH. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 20.Florea AM, Büsselberg D. Cisplatin as an anti-tumor drug: Cellular mechanisms of activity, drug resistance and induced side effects. Cancers (Basel) 2011;3:1351–1371. doi: 10.3390/cancers3011351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Zhang W, Guo X, Ren J, Gao A. The crosstalk between autophagy and apoptosis was mediated by phosphorylation of Bcl-2 and beclin1 in benzene-induced hematotoxicity. Cell Death Dis. 2019;10:772. doi: 10.1038/s41419-019-2004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daugas E, Susin SA, Zamzami N, Ferri KF, Irinopoulou T, Larochette N, Prévost MC, Leber B, Andrews D, Penninger J, Kroemer G. Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J. 2000;14:729–739. doi: 10.1096/fasebj.14.5.729. [DOI] [PubMed] [Google Scholar]
- 23.Koo MS, Kwon YG, Park JH, Choi WJ, Billiar TR, Kim YM. Signaling and function of caspase and c-Jun N-terminal kinase in cisplatin-induced apoptosis. Mol Cells. 2002;13:194–201. [PubMed] [Google Scholar]
- 24.Lin Y, Jiang M, Chen W, Zhao T, Wei Y. Cancer and ER stress: Mutual crosstalk between autophagy, oxidative stress and inflammatory response. Biomed Pharmacother. 2019;118:109249. doi: 10.1016/j.biopha.2019.109249. [DOI] [PubMed] [Google Scholar]
- 25.Bertero E, Maack C. Calcium signaling and reactive oxygen species in mitochondria. Circ Res. 2018;122:1460–1478. doi: 10.1161/CIRCRESAHA.118.310082. [DOI] [PubMed] [Google Scholar]
- 26.Sukumaran P, Nascimento Da Conceicao V, Sun Y, Ahamad N, Saraiva LR, Selvaraj S, Singh BB. Calcium signaling regulates autophagy and apoptosis. Cells. 2021;10:2125. doi: 10.3390/cells10082125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertero E, Maack C. Calcium signaling and reactive oxygen species in mitochondria. Circ Res. 2018;122:1460–1478. doi: 10.1161/CIRCRESAHA.118.310082. [DOI] [PubMed] [Google Scholar]
- 28.Carreras-Sureda A, Pihán P, Hetz C. Calcium signaling at the endoplasmic reticulum: Fine-tuning stress responses. Cell Calcium. 2018;70 doi: 10.1016/j.ceca.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Huang R, Hui Z, Wei S, Li D, Li W, Daping W, Alahdal M. IRE1 signaling regulates chondrocyte apoptosis and death fate in the osteoarthritis. J Cell Physiol. 2022;237:118–127. doi: 10.1002/jcp.30537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu H, Tian M, Ding C, Yu S. The C/EBP homologous protein (CHOP) transcription factor functions in endoplasmic reticulum stress-induced apoptosis and microbial infection. Front Immunol. 2019;9:3083. doi: 10.3389/fimmu.2018.03083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang Q, Li F, Shi K, Wu P, An J, Yang Y, Xu C. Involvement of p38 in signal switching from autophagy to apoptosis via the PERK/eIF2α/ATF4 axis in selenite-treated NB4 cells. Cell Death Dis. 2014;5:e1270. doi: 10.1038/cddis.2014.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lumley EC, Osborn AR, Scott JE, Scholl AG, Mercado V, McMahan YT, Coffman ZG, Brewster JL. Moderate endoplasmic reticulum stress activates a PERK and p38-dependent apoptosis. Cell Stress Chaperones. 2017;22:43–54. doi: 10.1007/s12192-016-0740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Y, Sun Q, Yuan F, Dong H, Zhang H, Geng R, Qi Y, Xiong X, Chen Q, Liu B. RND2 attenuates apoptosis and autophagy in glioblastoma cells by targeting the p38 MAPK signalling pathway. J Exp Clin Cancer Res. 2020;39:174. doi: 10.1186/s13046-020-01671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koromilas AE. M(en)TORship lessons on life and death by the integrated stress response. Biochim Biophys Acta Gen Subj. 2019;1863:644–649. doi: 10.1016/j.bbagen.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 35.Pljesa-Ercegovac M, Savic-Radojevic A, Matic M, Coric V, Djukic T, Radic T, Simic T. Glutathione transferases: Potential targets to overcome chemoresistance in solid tumors. Int J Mol Sci. 2018;19:3785. doi: 10.3390/ijms19123785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo C, Fordjour FK, Tsai SJ, Morrell JC, Gould SJ. Choice of selectable marker affects recombinant protein expression in cells and exosomes. J Biol Chem. 2021;297:100838. doi: 10.1016/j.jbc.2021.100838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang W, Yan Y, Chen M, Luo G, Hao J, Pan J, Hu S, Guo P, Li W, Wang R, et al. Aspirin enhances the sensitivity of colon cancer cells to cisplatin by abrogating the binding of NF-κB to the COX-2 promoter. Aging (Albany NY) 2020;12:611–627. doi: 10.18632/aging.102644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleih M, Böpple K, Dong M, Gaißler A, Heine S, Olayioye MA, Aulitzky WE, Essmann F. Direct impact of cisplatin on mitochondria induces ROS production that dictates cell fate of ovarian cancer cells. Cell Death Dis. 2019;10:851. doi: 10.1038/s41419-019-2081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Wang Z, Cai G, Huang P. Downregulation of circ_0071589 suppresses cisplatin resistance in colorectal cancer by regulating the MiR-526b-3p/KLF12 axis. Cancer Manag Res. 2021;13:2717–2731. doi: 10.2147/CMAR.S294880. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Sun M, He L, Fan Z, Tang R, Du J. Effective treatment of drug-resistant lung cancer via a nanogel capable of reactivating cisplatin and enhancing early apoptosis. Biomaterials. 2020;257:120252. doi: 10.1016/j.biomaterials.2020.120252. [DOI] [PubMed] [Google Scholar]
- 41.Sun X, Wang S, Gai J, Guan J, Li J, Li Y, Zhao J, Zhao C, Fu L, Li Q. SIRT5 promotes cisplatin resistance in ovarian cancer by suppressing DNA damage in a ROS-dependent manner via regulation of the Nrf2/HO-1 pathway. Front Oncol. 2019;9:754. doi: 10.3389/fonc.2019.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang YK, Wang YJ, Lei ZN, Zhang GN, Zhang XY, Wang DS, Al Rihani SB, Shukla S, Ambudkar SV, Kaddoumi A, et al. Regorafenib antagonizes BCRP-mediated multidrug resistance in colon cancer. Cancer Lett. 2019;442:104–112. doi: 10.1016/j.canlet.2018.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uozaki H, Horiuchi H, Ishida T, Iijima T, Imamura T, Machinami R. Overexpression of resistance-related proteins (metallothioneins, glutathione-S-transferase pi, heat shock protein 27, and lung resistance-related protein) in osteosarcoma: Relationship with poor prognosis. Cancer. 1997;79:2336–2344. doi: 10.1002/(SICI)1097-0142(19970615)79:12<2336::AID-CNCR7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 44.Singh RR, Reindl KM. Glutathione S-transferases in cancer. Antioxidants (Basel) 2021;10:701. doi: 10.3390/antiox10050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Y, Chang C, Huang R, Liu B, Bao L, Liu W. AP1 is essential for generation of autophagosomes from the trans-Golgi network. J Cell Sci. 2012;125:1706–1715. doi: 10.1242/jcs.093203. [DOI] [PubMed] [Google Scholar]
- 46.Chen P, Liu XQ, Lin X, Gao LY, Zhang S, Huang X. Targeting YTHDF1 effectively re-sensitizes cisplatin-resistant colon cancer cells by modulating GLS-mediated glutamine metabolism. Mol Ther Oncolytics. 2021;20:228–239. doi: 10.1016/j.omto.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong X, Yang Y, Zhou Y, Bi X, Zhao N, Zhang Z, Li L, Hang Q, Zhang R, Chen D, et al. Glutathione S-transferases P1 protects breast cancer cell from adriamycin-induced cell death through promoting autophagy. Cell Death Differ. 2019;26:2086–2099. doi: 10.1038/s41418-019-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abbasi A, Forsberg K, Bischof F. The role of the ubiquitin-editing enzyme A20 in diseases of the central nervous system and other pathological processes. Front Mol Neurosci. 2015;8:21. doi: 10.3389/fnmol.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das T, Chen Z, Hendriks RW, Kool M. A20/tumor necrosis factor α-induced protein 3 in immune cells controls development of autoinflammation and autoimmunity: Lessons from mouse models. Front Immunol. 2018;9:104. doi: 10.3389/fimmu.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang C, Zang W, Tang Z, Ji Y, Xu R, Yang Y, Luo A, Hu B, Zhang Z, Liu Z, Zheng X. A20/TNFAIP3 regulates the DNA damage response and mediates tumor cell resistance to DNA-damaging therapy. Cancer Res. 2018;78:1069–1082. doi: 10.1158/0008-5472.CAN-17-2143. [DOI] [PubMed] [Google Scholar]
- 51.Vendrell JA, Ghayad S, Ben-Larbi S, Dumontet C, Mechti N, Cohen PA. A20/TNFAIP3, a new estrogen-regulated gene that confers tamoxifen resistance in breast cancer cells. Oncogene. 2007;26:4656–4667. doi: 10.1038/sj.onc.1210269. [DOI] [PubMed] [Google Scholar]
- 52.Matsuzawa Y, Oshima S, Takahara M, Maeyashiki C, Nemoto Y, Kobayashi M, Nibe Y, Nozaki K, Nagaishi T, Okamoto R, et al. TNFAIP3 promotes survival of CD4 T cells by restricting MTOR and promoting autophagy. Autophagy. 2015;11:1052–1062. doi: 10.1080/15548627.2015.1055439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.