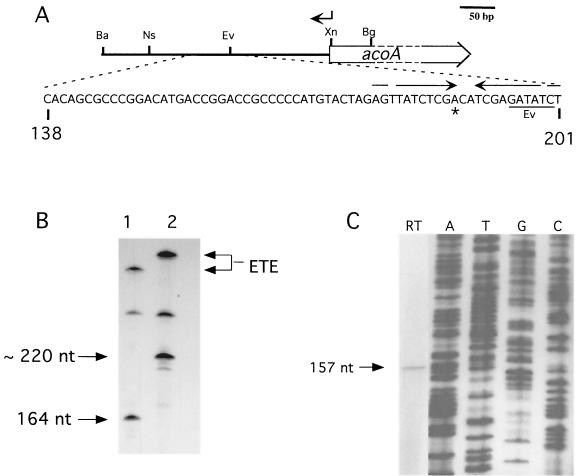
FIG. 6.
Mapping the acoA transcriptional start site. (A) The acoA transcriptional start site locus. Nucleotide coordinates refer to the acoA locus sequence. The location of the oligonucleotide used in primer extension is shown by an arrow above the map. Restriction sites: Ba, BamHI; Bg, BglII; Ev, EcoRV; Ns, NsiI; Xn, XmnI. The transcriptional start site (asterisk) mapped in vitro (B) and in vivo (C) was located in a region containing an inverted repeat (opposing arrows). (B) In vitro transcription using two different acoA promoter fragments and S. coelicolor RNAP. Runoff transcription was carried out using either the 348-bp BamHI/XmnI fragment (lane 1) or the 398-bp BamHI/BglII fragment (lane 2). The lengths of the two template fragments differed at the acoA proximal end, thus fixing the orientation of the runoff transcript. A larger, undefined runoff signal was also obtained from a promoter oriented oppositely to PacoA. The sizes of the runoff transcripts (arrows on the left) were estimated by separating the samples on a 6% polyacrylamide–6 M urea gel alongside a 32P-labeled M13mp18 sequencing ladder generated with a standard universal primer (5′-GCCAGGGTTTTCCCAGTCACGACG-3′). The signals generated by “end-to-end” (ETE) transcription are indicated on the right. (C) Reverse transcription assay (lane RT) using RNA isolated from exponential-phase S. coelicolor grown in YEME. An oligonucleotide overlapping with the acoA ORF (coordinates 321 to 344; indicated above by arrow; RT-Aco1 [5′-GACACGACAGTCTCCTTCATTTAT-3′]) was used as the primer. The size of the cDNA transcript (arrow) was estimated by running the sample alongside a 32P-labeled M13mp18 sequencing ladder generated with a standard universal sequencing primer (5′-TGTAAAACGACGGCCAGT-3′; lanes A, T, G, and C).