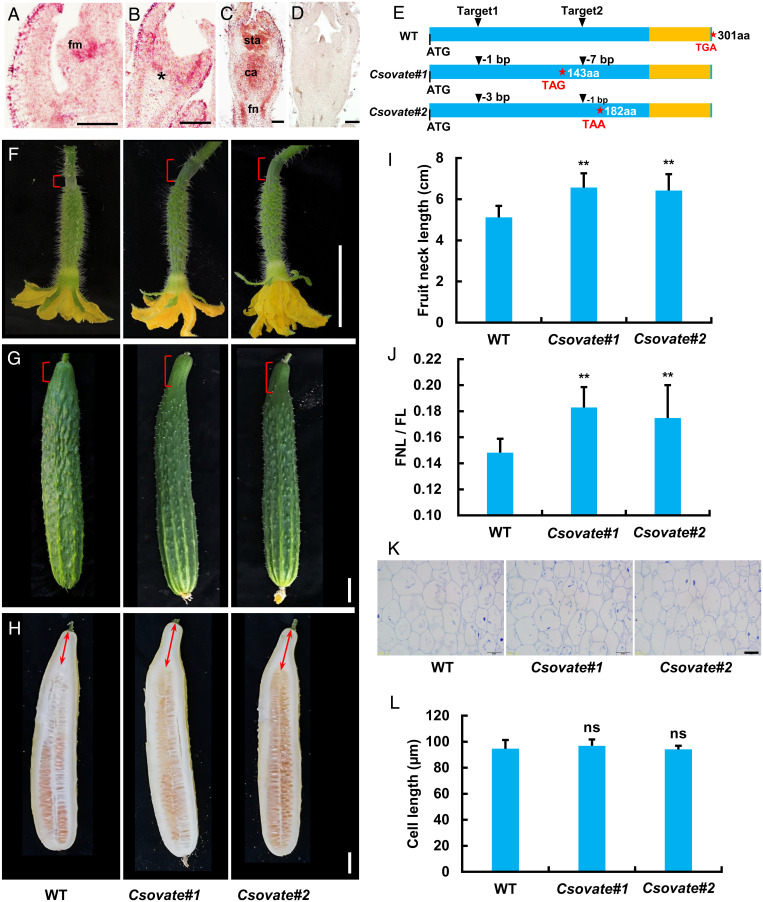
Fig. 4.
CsOVATE negatively regulates fruit neck elongation in cucumber. (A–D) In situ hybridization analysis of CsOVATE expression in cucumber. Longitudinal sections showing floral meristem (A) and developing flower primordia at stage 2 (B) and stage 6 (C). The asterisk indicates the developing fruit neck in cucumber. The negative control hybridized with the sense CsOVATE probe (D). (Scale bars, 100 µm.) (E) CRISPR-Cas9–induced homozygous mutations in CsOVATE producing knockout alleles due to early appearance of stop codons. The stars represent the termination codon position and the yellow boxes indicate the OVATE domain. (F–H) Fruit morphology of WT and Csovate mutants at 0 DPP (F), 10 DPP (G), and 40 DPP (H). The brackets indicate the fruit neck and the double arrows represent the measured FNL. (Scale bars, 2 cm.) (I and J) Quantification of FNL (I) and ratio of FNL/FL (J) in WT and Csovate mutants at 40 DPP. Values are means ± SD (n = 9 to 14). (K and L) Representative cell morphology (K) and cell-length quantification (L) in the longitudinal sections of fruit necks in WT and Csovate fruits at 40 DPP. Values are means ± SD (n = 9). (Scale bar, 50 µm.) Significance analysis compared with WT was performed with the two-tailed Student’s t test (ns, no significant difference; **P < 0.01).