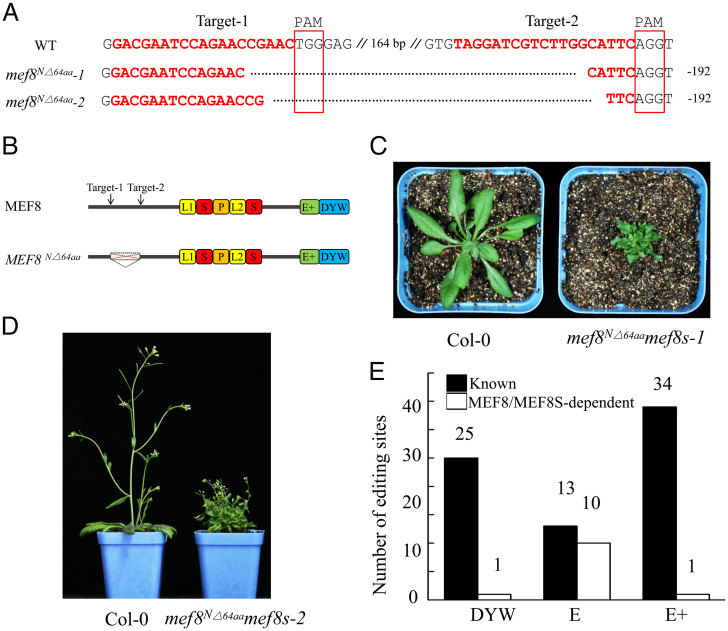
Fig. 3.
MEF8 and MEF8S are required for the editing of the E-type PPR-targeted sites in mitochondria. (A) Alignment of sequences of mutated alleles generated by CRISPR-Cas9. The fragments between two target sites were deleted. PAM, protospacer adjacent motif; WT, wild type. (B) Schematic illustration of MEF8 protein. The deleted region is indicated. (C and D) Morphology of the wild type and the mef8N△64aamef8s mutants. The seeds were germinated on MS media, and 1-wk-old seedlings were transferred to soil and grown in the greenhouse under long-day conditions (16 h light/8 h darkness). Images of a 4-wk-old wild-type plant and 6-wk-old mef8N△64aamef8s-1 mutant (C) and 6-wk-old wild-type plant and 10-wk-old mef8N△64aamef8s-2 mutant (D) were taken. The mef8N△64aamef8s mutants were slow-growing and produced sterile flowers. (E) RNA-editing analysis of the wild type and the mef8N△64aamef8s mutants. The editing analysis of 4-wk-old seedlings of the wild type and two independent lines of mef8N△64aamef8s mutants is shown. The editing status of the wild type and two independent lines of mef8N△64aamef8s mutants in mitochondria was analyzed by STS-PCRseq and Sanger sequencing of PCR products, respectively. If the editing extent of one site in the mef8N△64aamef8s mutant is decreased over 20% of that in the wild type, this site is considered as depending on MEF8/MEF8S. The known PPRs associated with editing sites are listed in Dataset S1B according to the literature (18, 19).