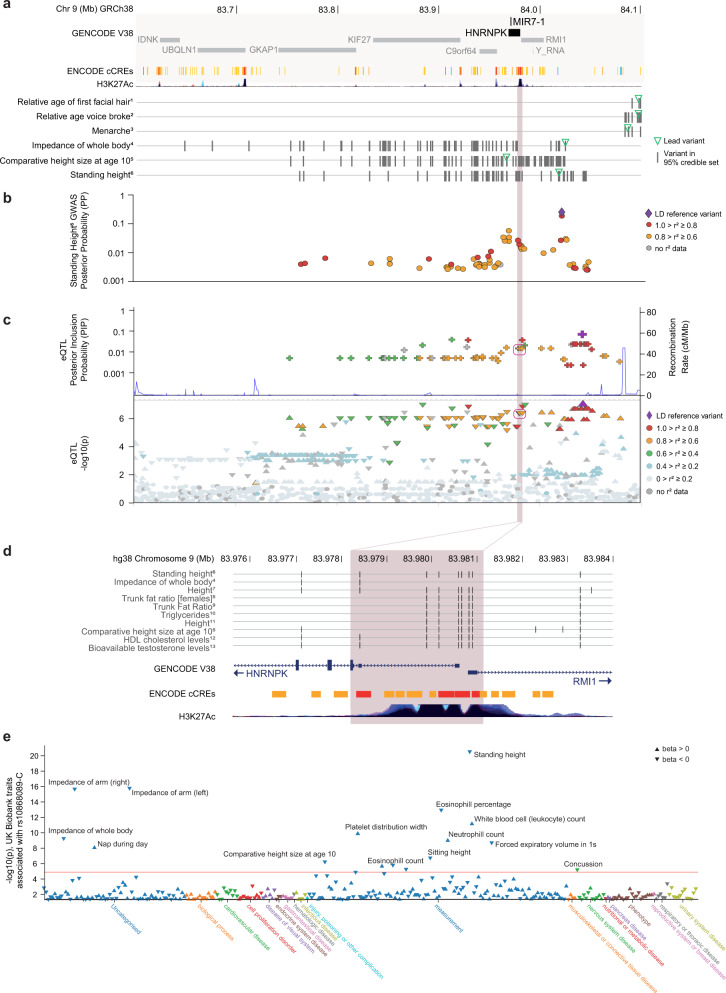
Fig. 6. Association of human variants in the locus encompassing MIR7-1 with height, adiposity and related traits.
a MIR7-1 is located in the last intron of HNRNPK (dark grey, GENCODE V38; chr9:83.6-84.1 Mb). Tracks display ENCODE candidate cis-Regulatory Elements (ENCODE cCREs112; red, promotor-like; orange, proximal enhancer-like; yellow: distal enhancer-like; blue: CTCF-only) and histone acetylation (H3K27Ac) tracks (UCSC browser hg38, (http://genome.ucsc.edu113). Tracks labelled with phenotypic traits (UK Biobank Neale v2, 2018) show the position of fine-mapped variants with Open Targets variant-to-gene (V2G) annotations for HNRNPK within this chromosomal region. Trait superscripts 1-6 refer to study-locus summary statistics in Supplementary Data 1b. Summary statistics for study-locus associations which had an Open Targets locus-to-gene (L2G) annotation for HNRNPK are provided in Supplementary Data 1. b Fine-mapping of GWAS locus for standing height, shown as posterior probability (PP) for each variant in the 95% credible set (UK Biobank-derived trait NEALE2_50_raw, lead variant chr9:84,020,284 G > C; data from Open Targets; Supplementary Data 1). The shaded area indicates a region where credible variants overlap with predicted regulatory regions immediately 5’ of HNRNPK. Six of these seven variants are in high pairwise LD (r2 > 0.95). c LocusZoom of the reported cis-eQTL for HNRNPK expression in monocytes (BLUEPRINT); data from FIVEx, https://fivex.sph.umich.edu. Open Targets reported evidence of colocalisation with standing height (UKB Neale v2, 2018; coloc114, H4 = 0.81, QTL beta = 0.04). eQTL minimum p-value was at rs10868089-C (chr9:84,042,868 T > C, LD reference variant). d Zoomed-in view showing predicted ENCODE cis-Regulatory Elements immediately 5’ of HNRNPK (chr9:83.976-83.984 Mb). Displays variants which are contained in a credible set of a selected trait (for UK Biobank-derived traits) or are in linkage disequilibrium (r2 > 0.8) with the lead variant (for non-UK Biobank studies). Trait superscripts 4-13 refer to study-locus summary statistics in Supplementary Data 1b. e Single-variant summary statistics (p-value and direction of effect) for the HNRNPK eQTL lead variant shown in (c) (rs10868089-C, chr9:84042868 T > C) obtained from Open Targets. Points represent UK Biobank-derived traits (UK Biobank Neale v2, 2018) with single-variant P < 0.05.