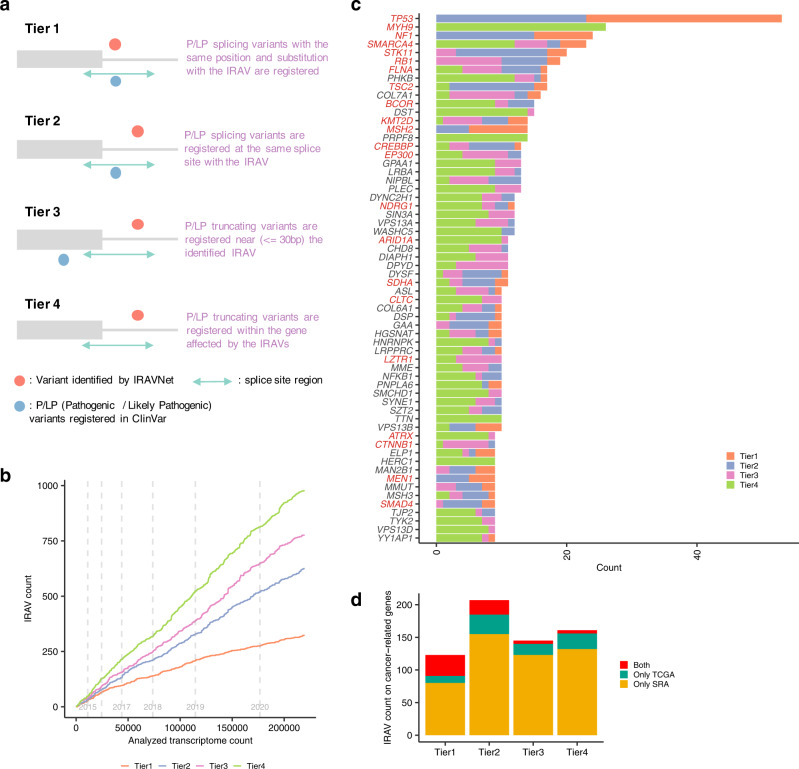
Fig. 4. The landscape of putative pathogenic IRAVs.
a A conceptual diagram for the classification of disease-related IRAVs in four tiers. The ranks of putative pathogenic IRAVs are determined by comparison with the positional relationships with the registered pathogenic/likely pathogenic (P/LP) variants in ClinVar. b The saturation analysis of putative pathogenic IRAVs via using Sequencing Read Archive. Transcriptome data were sorted by registration date, and the cumulative number of putative pathogenic IRAVs against the number of the analyzed transcriptome is shown. Gray vertical lines indicate the registration year changes. c The landscape of putative pathogenic IRAVs for frequently altered genes (total number ≥10). Genes were sorted by the total number of distinct variants, and known cancer-related genes are shown in red. d The number of distinct putative pathogenic IRAVs affecting cancer-related genes for each rank identified from Sequencing Read Archive, TCGA, and both.