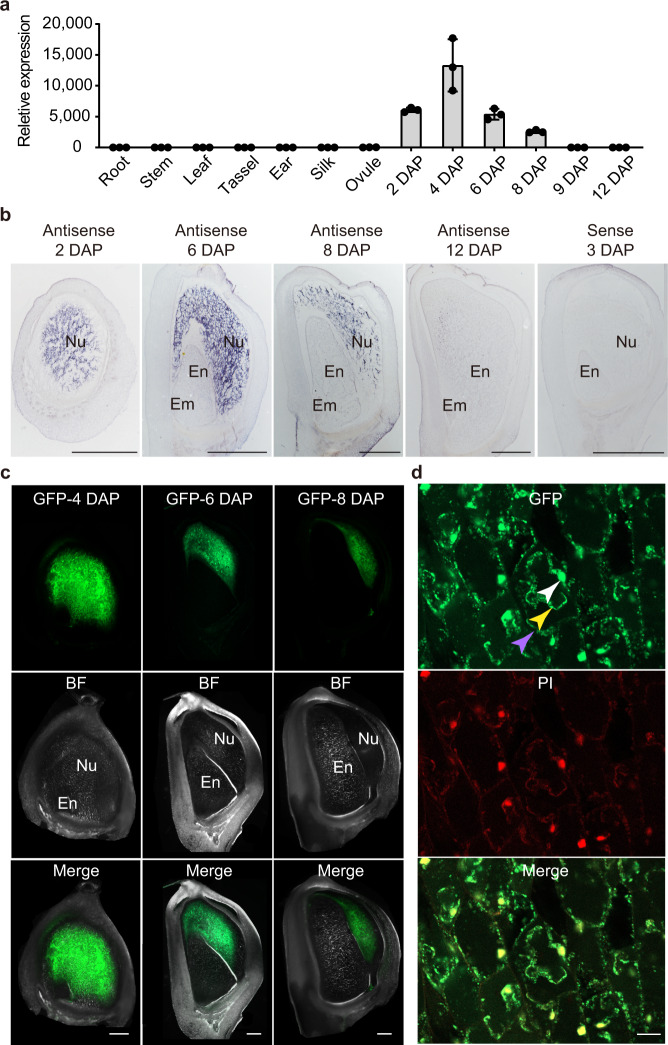
Fig. 3. ZmEXPB15 accumulates specifically in nucellus of developing kernels.
a qRT-PCR analysis of ZmEXPB15 in root, stem, leaf, tassel, ear, silk and developing kernels at 0 (ovule) to 12 DAP. All expression levels from three biological repeats were normalized to Actin. Values are means ± s.d. b In situ localization of ZmEXPB15 mRNA in developing kernels at 2 to 12 DAP. Positive signals (shown in purple) are clearly restricted to the nucellus at 2 to 8 DAP (first three panels) using the ZmEXPB15 antisense probe. No signal is observed in the section of a 12-DAP kernel (fourth panel). The control was performed using 3-DAP kernel with a ZmEXPB15 sense probe (last panel). c Localization of ZmEXPB15-GFP in developing kernels at 4, 6 and 8 DAP from proZmEXPB15:ZmEXPB15-GFP transgenic plants. The ZmEXPB15-GFP signal was specifically detected in the nucellus. BF bright-field image, Merge merge of GFP and BF images. Nu nucellus, En endosperm, Em embryo. d ZmEXPB15-GFP expression was observed in the cell wall (purple arrow), cytoplasm (yellow arrow) and nucleus (white arrow) of the nucellar cells in 4-DAP kernels. PI, propidium iodide staining. Merge, merge of GFP and PI images. The experiments in (b–d) were repeated two times with a similar result. Scale bar = 1 mm in (b); 500 µm in (c); 20 µm in (d). Source data are provided as a Source Data file.