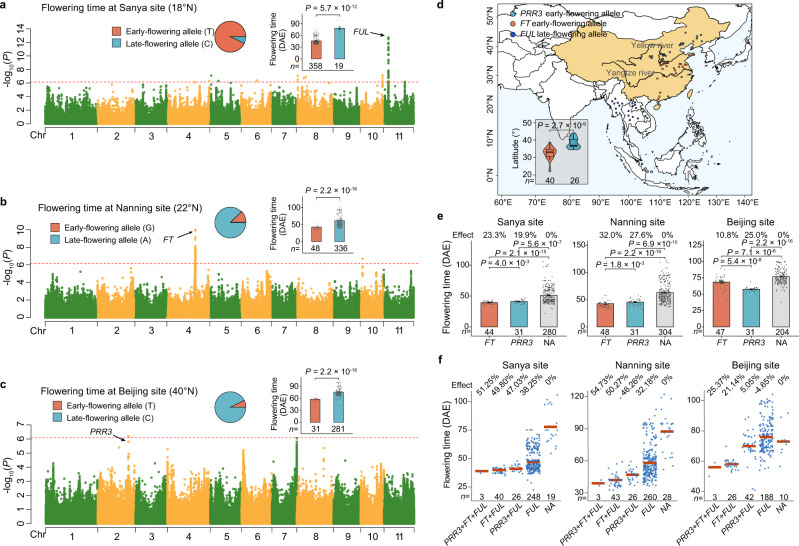
Fig. 4. The genetic architecture underlying flowering time control from low to high latitudes.
a–c Manhattan plots of GWAS for flowering-time data measured in Sanya (18°N) in 2021 (a), Nanning (22° N) in 2020 (b), and Beijing (40°N) in 2021 (c). Red horizontal dashed line indicated the Bonferroni-corrected significance thresholds of GWAS (α = 1). Pie charts represented allelic frequencies of the major associated loci. The bar plots display the flowering time of landraces carrying each allele of the identified major loci. DAE, days after emergence. The number (n) of landraces carrying each allele is shown below. d The geographical distributions of landraces carrying the early-flowering alleles of FT and PRR3, and the late-flowering allele of FUL, respectively. The map was created using the map_data() function in the R package ggplot2. The violin plot showed significant differences in latitudes between landraces carrying the early-flowering allele of FT (n = 40) and PRR3 (n = 26), respectively. In the box plots, central line: median values; bounds of the box: 25th and 75th percentiles; whiskers: 1.5*IQR (IQR: the interquartile range between the 25th and 75th percentile). e The bar plots show the flowering shortening effects of the early-flowering alleles of FT and PRR3 at each of the three measurement sites. NA indicates landraces carrying neither of these two early-flowering alleles. The number (n) of landraces carrying each allele at each of the three measurement sites is shown below. f The dot plots show the flowering time shortening effects of early-flowering allelic combinations at each of the three measurement sites. Blue dots represent the landraces categorized according to all of the different allelic combinations found in the 440 sequenced landraces. Red lines indicate the average value of each category. NA indicates landraces carrying no early-flowering alleles. The number of landraces for each category is shown below. The significance was tested with two-sided Wilcoxon tests in (a–e). The data in a–c and e are shown as mean ± SE, and the error bars represent SE. Source data are provided as a Source Data file.