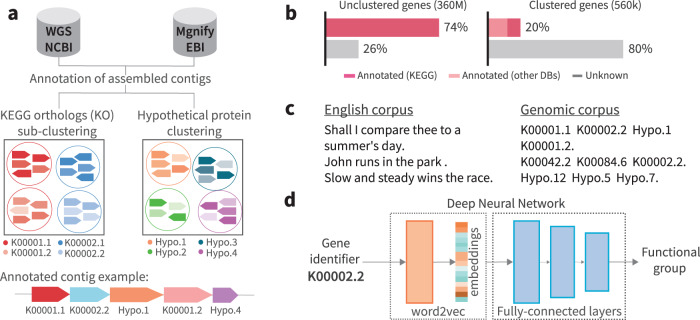
Fig. 1. Model workflow.
a Data processing and annotation. Assembled contigs from public databases were downloaded and underwent gene calling and annotation. Both annotated and unannotated genes were clustered into gene families. b Distribution of annotated and hypothetical genes in the corpus. The left bar chart represents all ~360 million genes used in the corpus. The right bar chart represents ~560,000 unique gene families (i.e., the corpus “vocabulary”). c A comparison between English and genomic corpora. The “sentences” in the genomic corpus are contigs, which are composed of gene families identifiers as “words”. d Embedding generation and function prediction pipeline. Embeddings (numeric vector representations) are generated by the word2vec algorithm and serve as the input to a deep neural network for gene function classification.