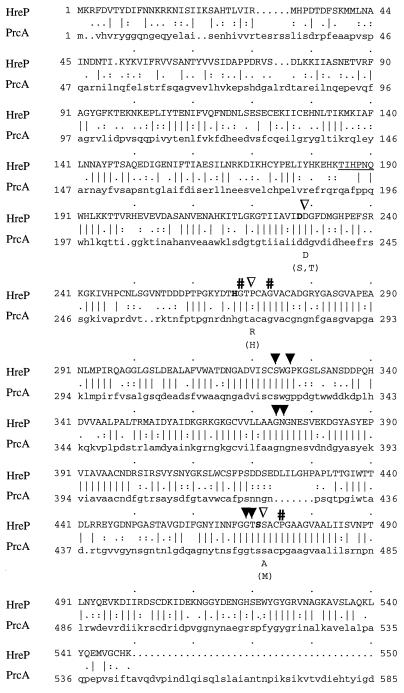
FIG. 1.
Amino acid alignment of Y. enterocolitica HreP and A. variabilis PrcA. Identical residues are indicated by vertical lines, and similar residues are indicated by periods or colons (Bestfit; Genetics Computer Group). The amino acids that make up the typical catalytic triad of serine proteases are highlighted by boldface letters. Solid arrowheads, amino acids that are predicted to be located in the putative substrate binding region; open arrowheads, positions of amino acids typical for the kexin subfamily of subtilases; #, amino acids predicted to be located in internal helices. Those amino acids that are characteristic for kexin proteases or serine proteases (in brackets) are indicated under the alignment. The amino acids that were determined by N-terminal sequencing of the 42-kDa polypeptide are underlined and correspond to the amino-terminal end of the mature HreP protease.