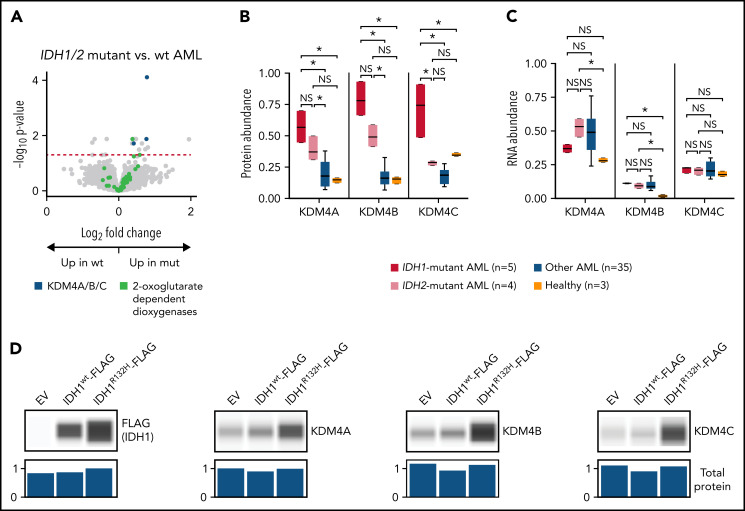
Figure 3.
AML samples with IDH1/2 mutations are associated with increased abundance of KDM4A/B/C histone demethylases. (A) Volcano plot showing protein abundance in IDH1/IDH2-mutated vs wt AML samples. P values are calculated using the t test and corrected for multiple-hypothesis testing with the Benjamini-Hochberg method. Dashed red line shows P = .05. IDH1 and IDH2 mutations in AML cause dysregulation of 2-oxoglutarate metabolism, and KDM4A/B/C are some of the known 2-oxoglutarate–dependent dioxygenases. (B-C) Normalized abundance of KDM4A/B/C classified by IDH1/2 mutation status in TMT data (B) and bulk RNA sequencing (C). *P < .05 by t test between groups. (D) K562 cells were transfected in vitro with pcDNA3-EV, pcDNA3-FLAG-IDH1wt, or pcDNA3-FLAG-IDH1R132H plasmids and cultured for 2 days prior to cell lysis. Western blots show 1 of 3 representative biologic replicates using antibodies specific for the indicated proteins. Normalized total protein values as a loading control were calculated using the protein normalization module on the Jess western blotting system. EV, empty vector; mut, mutant; NS, not significant.