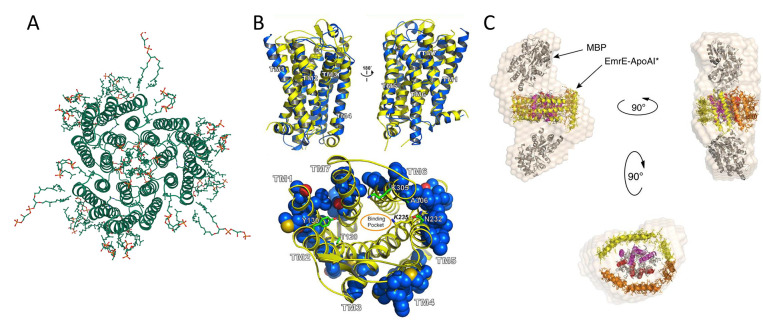
Figure 11.
Redesign of multipass transmembrane receptors. (A) Natural trimer model basis for the redesign of soluble bacteriorhodopsin (PDB ID: 2BRD, created by Mol*294). (B) Top: structure superimposition of murine MUR (PDB ID: 4DKL, yellow color) and wsMUR-TM receptor (blue color). Bottom: mutated positions in wsMUR are depicted as blue spheres, the majority of which (50 out of 55) are located at the exterior of the structure. Reprinted with permission from ref (262). Copyright 2013 Perez-Aguilar et al. (C) Multiple views of particle envelope reconstruction calculated ab initio on ΔspMal-bp-EmrE-ApoAI* from SAXS data. ΔspMal-bp crystal structure (PDB ID: 1NL5); ApoAI lipid-free crystal structure (PDB ID: 2A01); and electron microscopy-derived structure of dimeric EmrE (PDB ID: 2I68). The MBP label in the graph refers to maltose-binding protein (Mal-bp) defined in the manuscript. Reprinted with permission from ref (267). Copyright 2015 Mizrachi et al.