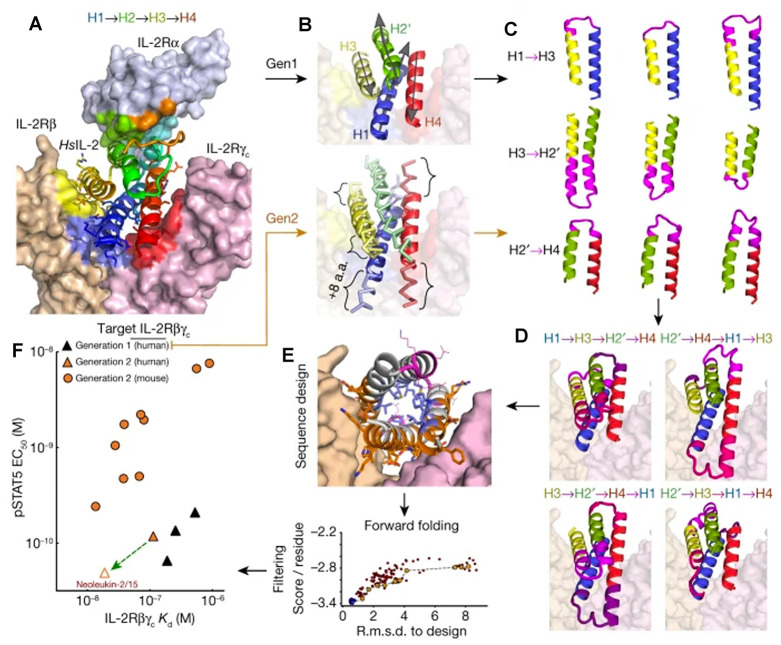
Figure 21.
(A) Structure of human IL-2 (HsIL-2 in the graph) in complex with its receptor IL-2Rαβγc (surface representation) (PDB ID: 2B5I). (B) Designed mimics have four helices; three (blue, yellow, and red) mimic IL-2 interactions with IL-2Rβγc, whereas the fourth (green) holds the first three in place. Top, first iteration: each of the core elements of IL-2 (helices H1–H4) were independently idealized by the assembly of four residue clustered protein fragments. Bottom, second iteration: the core elements were built using parametric equations that recapitulate the shape of each disembodied helix, allowing changes in the length of each helix by up to ±8 amino acids. (C) Pairs of helices were reconnected using ideal loop fragments. (D) Combinations of helix hairpins in C to generate fully connected protein backbones. (E) Rosetta flexible backbone sequence design. (F) Binding and activity of selected designs (solid symbols). The green arrow originates at the parent of the optimized design Neo-2/15. Reprinted with permission from ref (528). Copyright 2019 Springer Nature.