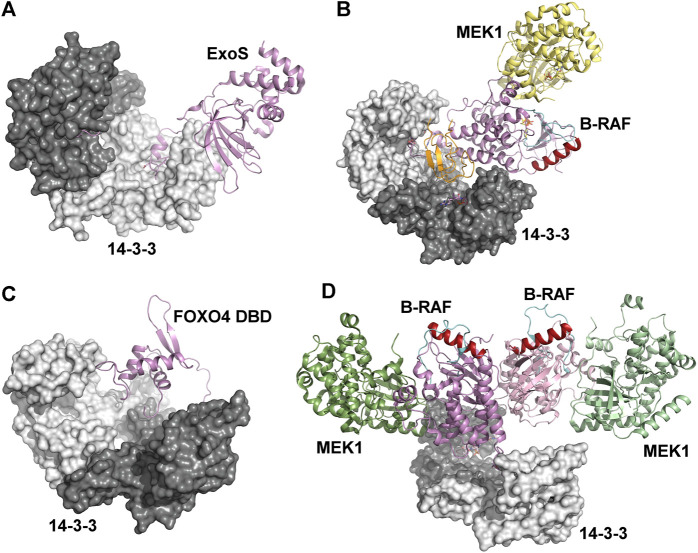
FIGURE 4.
Mechanisms based on physical occlusion and facilitation of protein-protein interactions. (A) Crystal structures of the ExoS:14-3-3β complex showing an extensive hydrophobic binding interface. The amphipathic 14-3-3 binding LDLA-motif is shown as sticks (Karlberg et al., 2018). (B) Autoinhibited B-RAF:MEK1:14-3-3 complex (PDB ID: 6NYB (Park et al., 2019)). The CRD and the catalytic domains of B-RAF are shown in orange and violet, respectively. The position of the C-helix (shown in dark red) and the activation segment (shown in cyan) correspond to the autoinhibited state. (C) The Förster resonance energy transfer-based model of the complex between the FOXO4 Forkhead domain (FOXO4 DBD) and 14-3-3ζ (Silhan et al., 2009). The 14-3-3 binding motifs is located at the C-terminus of FOXO4-DBD. (D) The active B-RAF:MEK1:14-3-3 complex (PDB ID: 6Q0J (Park et al., 2019)). The 14-3-3 dimer anchors C-terminal 14-3-3 binding motifs of two B-RAF molecules, and the B-RAF kinase domains are oriented in the back-to-back fashion with the C-helix (shown in dark red) in a position consistent with the active conformation. The figure was prepared with PyMOL (https://pymol.org/2/).