Figure 2. The NaVMs tamoxifen receptor site is conserved in domain II and IV of human NaV1.7.
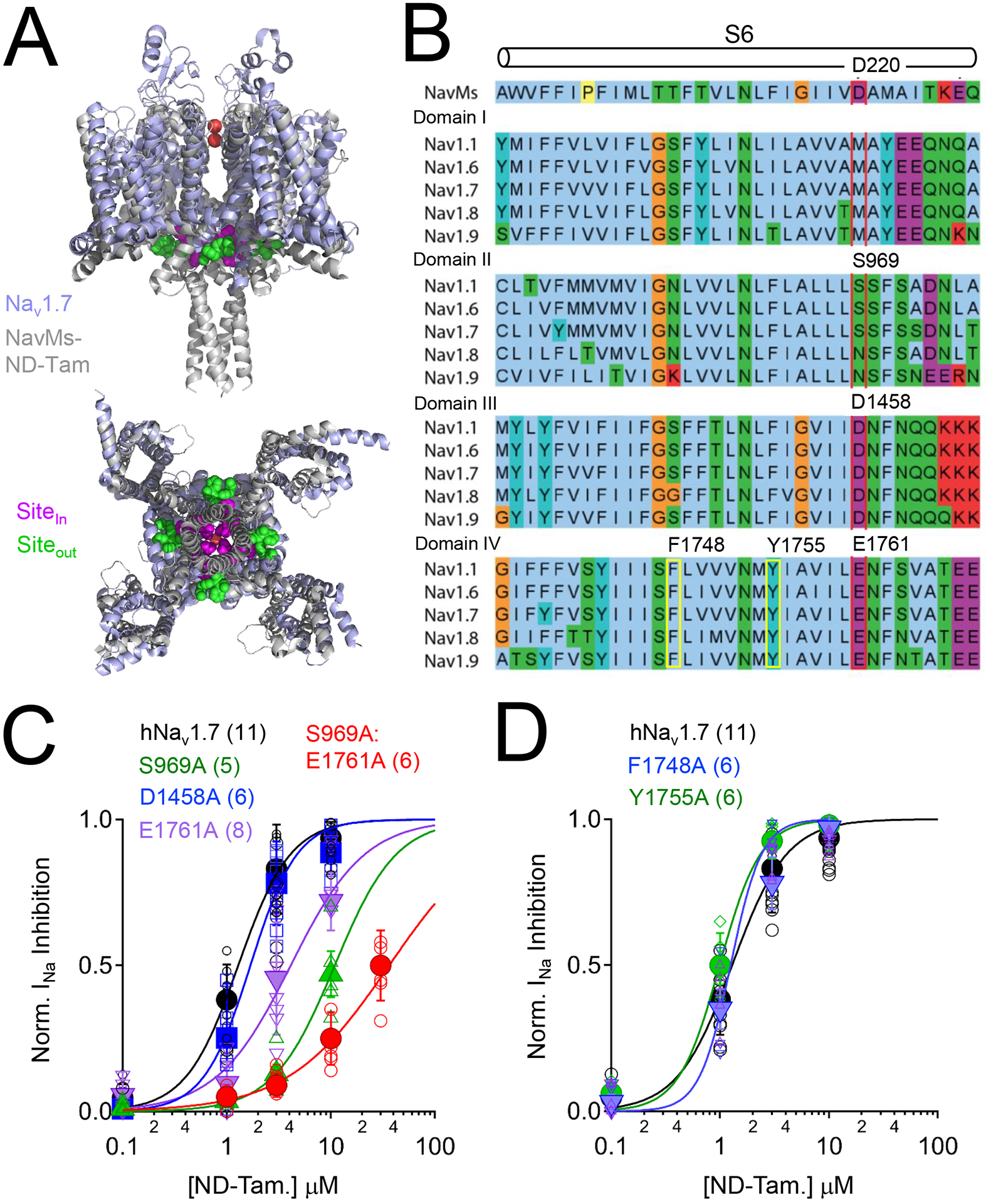
A) Structural alignment of human NaV1.7 (PDB: 6J8J) and the ND-Tam bound prokaryotic NavMs F208L channel (PDB: 6SXG)(Shen et al., 2019, Sula et al., 2021). The NavMs and NaV1.7 channel structures are reported to be in either pre-open or inactivated states. Inner (Sitein) and outer (Siteout) ND-Tam binding sites are indicated. B) Multiple sequence alignment of the sixth transmembrane segment (S6) from NavMs (UnitProt ID: A0L5S6) and each domain (I-IV) of DRG sodium channels NaV1.1 (Q99250), NaV1.6 (Q9UQD0); NaV1.7 (Q15858); NaV1.8 (Q9Y5Y9) and NaV1.9 (Q9UI33) using the standard Clustal color scheme by amino acid character. Proposed location of the ND-Tam receptor is boxed in red. The previously identified local antiesthetic/ anti-arrhythmic receptor site is boxed in yellow. C) Comparative ND-Tam potency for NaV1.7 channels expressing alanine substitutions at proposed receptors for tamoxifens, and D) local anesthetic receptor sites. Drug concentration- INa inhibition relationships for ND-Tam are fit to the Hill equation. Open symbols represent responses from individual cells and filled symbols represent average response per concentration. Error is equal to S.E.M. and the number of cells evaluated per treatment group is indicated within the parentheses.
