FIG. 1.
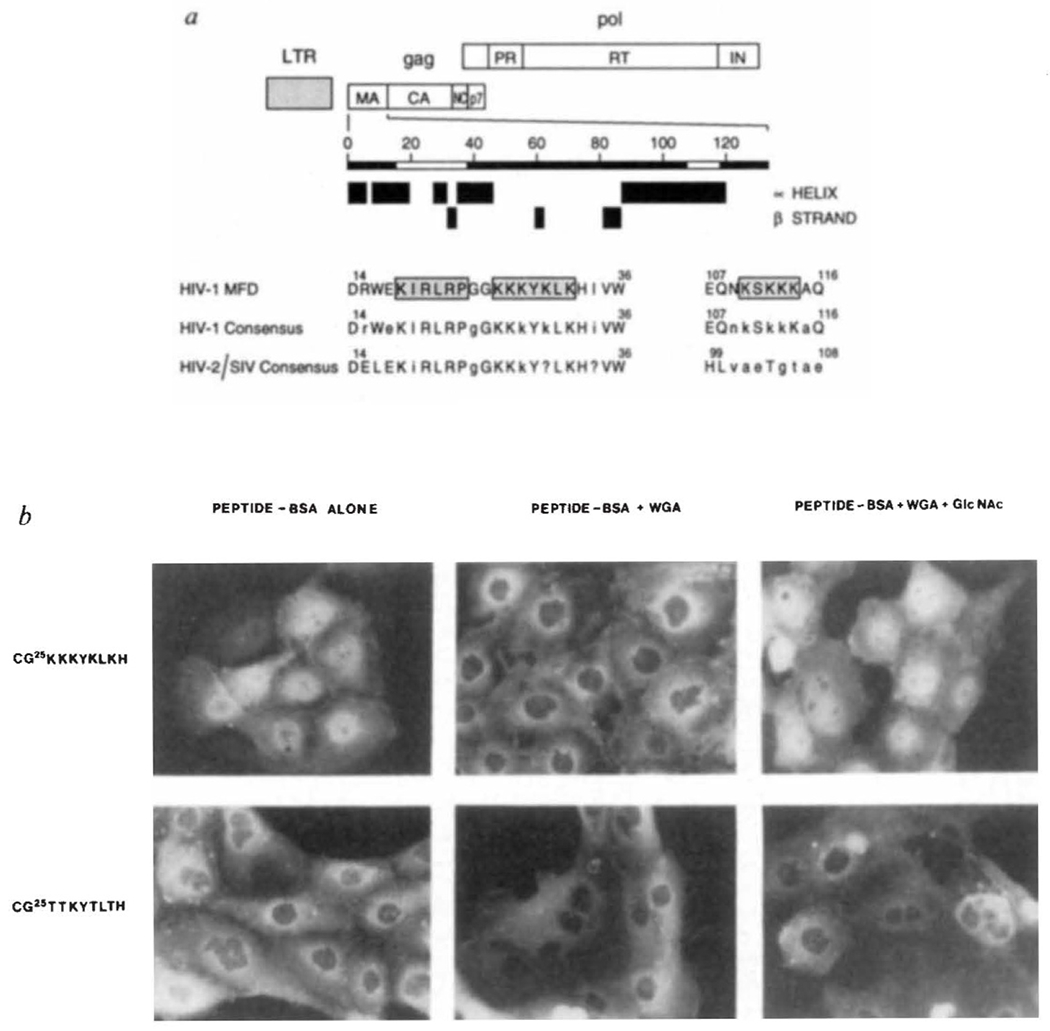
Putative nuclear targeting sequences within the HIV-1 gag matrix protein. a, Boundaries of the major products of the gag and pol polyproteins are indicated. The positions of predicted α-helices and β-strands were determined as in ref. 21. Sequences that resemble common NLS motifs6 are boxed and stippled. Amino-acid numbering, alignment and description of consensus sequences was helped by information contained within the HIV-1 sequence data base22. Upper case letters represent invariant residues whereas conserved and non-conserved amino acids are denoted by lower case letters and the question mark, respectively. b, Subcellular location of HIV-1 matrix NLS peptide–BSA conjugates after microinjection into rat epithelial cells.
METHODS. PtK1 cells (rat kangaroo kidney epithelia) were grown on glass cover slips in F-12 medium supplemented with 10% fetal bovine serum. Cells were microinjected in Hank’s balanced salt solution, essentially as detailed elsewhere8. Fluoresceinated HIV-1 matrix peptide–BSA conjugates containing an average of 6 peptides per BSA molecule7 were injected with or without WGA alone (2.5 mg ml−1) or WGA and its ligand, GlcNAc(0.5 M). Microinjected cells were incubated for 30 min at 37 °C, fixed and photographed8.
