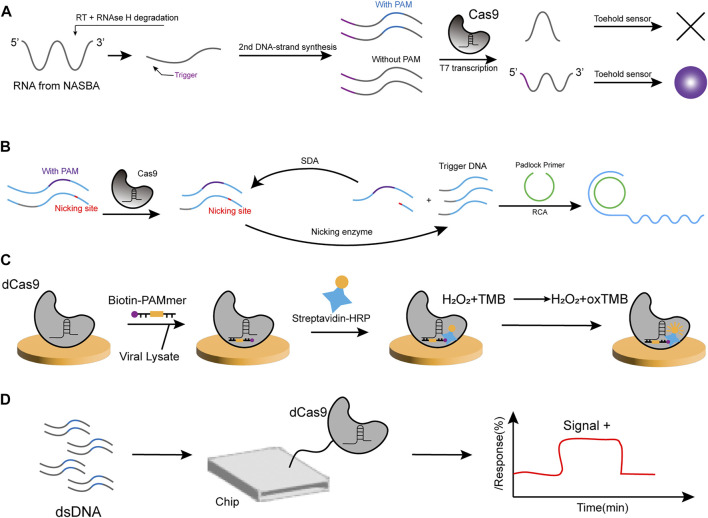
FIGURE 2.
Strategies for Cas9-based nucleic acid sensor. (A) NASBA amplifies the RNA target, and the trigger sequence for the toehold switch is introduced. Subsequently, the RNA and DNA hybridization produces RNA that is degraded by RNAse H. The second DNA strand is synthesized, and it carries the T7 promoter sequence. The transcribed RNA can be used as starting material for NASBA and interact with the toehold switch. In the presence of PAM sequences, the RNA target site is shorter due to the cleaved template and cannot activate the sensor, and vice versa, producing a significant color change. (B) In the action of Cas9, broken dsDNA at the nicking site is further recognized by the nicking enzyme. Once the DNA polymerase reaches the nicking site, a new strand can be generated and the downstream strand is replaced. As a result, many short ssDNAs are generated to trigger rolling circle amplification (RCA). Subsequently, the RCA product can bind to oligonucleotide-spliced AuNPs, inducing the aggregation of AuNPs. SDA, Strand-displacement amplification. (C) Viral lysate and biotin-PAMmer are added to microplates immobilized with dCas9/gRNA complexes. Following, Streptavidin-HPR and TMB substrate solutions were added to the microplate. Finally, the yellow color is observed in the presence of the virus. (D) The CRISPR-chip is composed of a gFET structure with a complex of sgRNA and dCas9 formed on the graphene surface. When the target DNA is detected, dCas9 binds to the target DNA, which modulates the electrical properties of the gFET and leads to an electrical signal output. gEFT: graphene-based field-effect transistor.