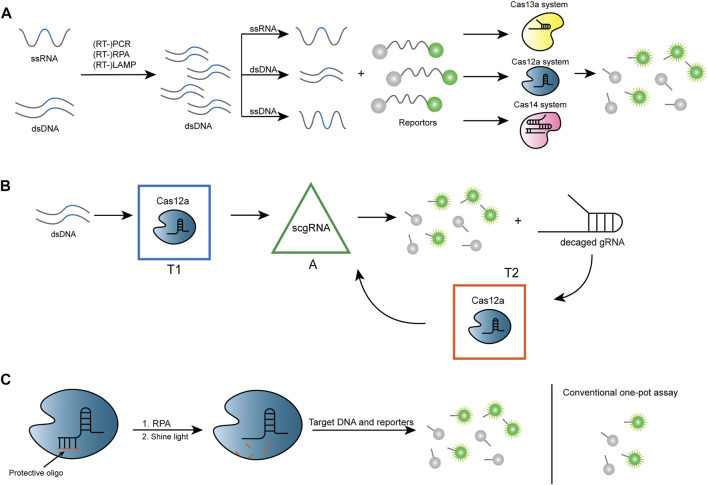
FIGURE 3.
Strategies for Cas12 and Cas13-based nucleic acid sensor. (A) After amplification, ssRNA or dsDNA forms dsDNA and produces ssRNA, dsDNA, and ssRNA, depending on the different systems. After being mixed with fluorescent reporters, trans cleavage occurs through specific binding of different CRISPR/Cas systems, which subsequently generates a fluorescent signal. (B) In CONAN, there are three signal processors, namely transducer 1 (T1), transducer 2 (T2), and signal amplifier (A), where the parallel connection of A and T2 forms a positive feedback circuit and T1 is connected in series with the positive feedback circuit. Cas12a and gRNA for target dsDNA (gRNA-T) are pre-assembled as T1, which converts the signal input into active Cas12a protein (Cas12a/gRNA-T/DNA complex). After the action of a self-reporting scgRNA, an amplified fluorescent signal and multiple active (decaged) gRNA molecules are output. T2 contains Cas12a protein and a probe for decaged gRNA, which can transduce the resulting decaged gRNA into another active Cas12a protein to produce an amplified fluorescent signal. scgRNA: switchable-caged guide RNA. (C) The protective oligonucleotide is designed to partially pair with the crRNA, thereby altering the conformation of the crRNA so that it cannot bind to the target DNA and inhibit the cleavage activity of Cas12a. The protective oligonucleotide breaks when exposed to ultraviolet (UV) light at 365 nm isolated from crRNA, thereby restoring the function of CRISPR-Cas12a detection. This system ensures that the target DNA is not reduced by Cas12a during the amplification phase, resulting in a stronger fluorescent signal.