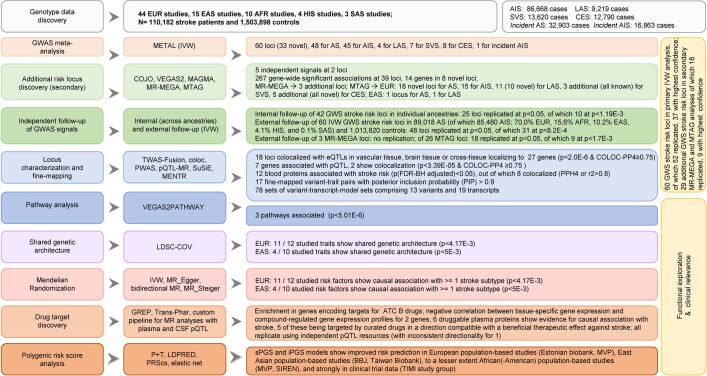
Extended Data Fig. 1. GIGASTROKE study workflow.
Study workflow and rationale. EUR: European; EAS: East-Asian; AFR: African; HIS: Hispanic; SAS: South Asian; AS: any stroke; AIS: any ischaemic stroke; LAS: large artery stroke; CES: cardioembolic stroke; SVS: small vessel stroke; GWAS: genome-wide association study; IVW: inverse-variance weighted; MR-MEGA: meta-regression of multi-ethnic genetic association; COJO:conditional and joint analysis; VEGAS2:versatile gene-based association study 2; MTAG: multi-trait analysis of GWAS; TWAS: Transcriptome-wide association study; coloc: Colocalisation Test; PWAS: Proteome-wide association studies; pQTL-MR: protein quantitative trait loci Mendelian Randomization; SuSiE: sum of single effects model; MENTR: Mutation Effect prediction on Non-coding RNA TRanscription; PIP: posterior probability; FDR: false discovery rate; LDSC-COV: covariate-adjusted LD score regression; MR-Egger: Mendelian randomization-Egger; GREP: genome for REPositioning drugs; ATC: Anatomical Therapeutic Chemical; P+T: pruning and thresholding; PRScs: polygenic risk score under continuous shrinkage; BBJ: Biobank Japan; TIMI: thrombolysis in myocardial infarction; MVP: Million Veteran Program; SIREN: Stroke Investigative Research and Educational Network.