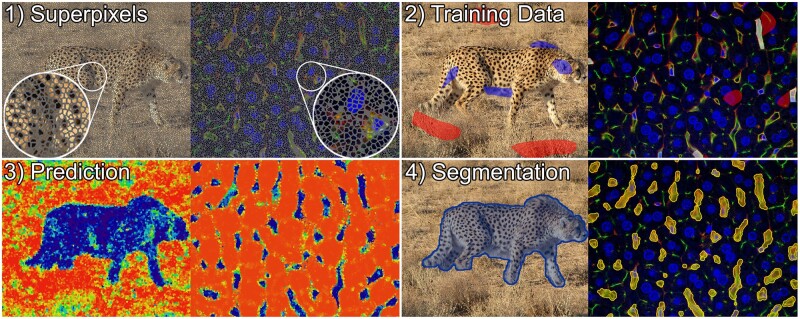
Fig. 2.
Illustration of supervoxel-based image segmentation procedure on an image of a cheetah (left) (photo by Behnam Ghorbani, published via Wikimedia Commons CC 4.0), and a 3D confocal micrograph of mouse liver tissue in which blood vessels were segmented (right). The capillaries in this imaging setup appear yellow–green and usually comprise an unstained lumen as well as few small, elongated nuclei colored blue, which typically belong to endothelial (sinusoidal) cells forming the capillary wall, immune cells residing in the lumen or other non-parenchymal cells. (1) Supervoxel outlines are shown in gray. (2) Training data for the background class is colored red in both instances, while the foreground class is colored blue in the left and white in the right example. Annotations were selected to represent the characteristic visual features of both classes. For example, annotations of capillaries comprise yellow-colored walls, lumen and enclosed cell nuclei, whereas the background annotations encompass unstained cytoplasm and nuclei of the parenchymal liver cells (blue), bile canaliculi (green) as well as regions close to capillaries to promote learning of exact boundaries. (3) Class membership probabilities in the prediction images are illustrated using a color mapping ranging from red (low probability of being foreground) over yellow to blue (high probability of being foreground). (4) The segmentation is visualized by a blue overlay on the left, and a yellow overlay on the right (A color version of this figure appears in the online version of this article.)