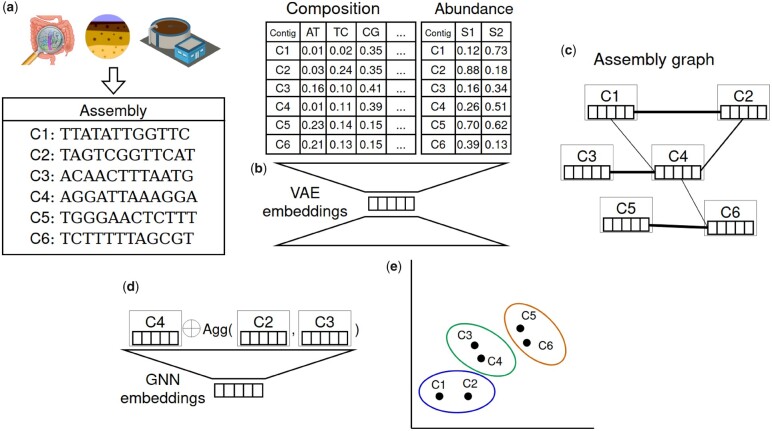
Fig. 1.
GraphMB’s workflow. (a) The metagenome of an environmental sample is sequenced and assembled into contigs. (b) Initial embeddings are computed with a variational auto-encoder based on k-mer composition and abundance features. (c) The input of the GNN are the initial contig embeddings and the graph structure provided by the assembly graph. The thickness of the edge corresponds to the number of reads that cover it. (d) The GNN model learns new embeddings by aggregating neighboring contigs (nodes in the assembly graph). (e) The final embeddings are clustered and bins are obtained