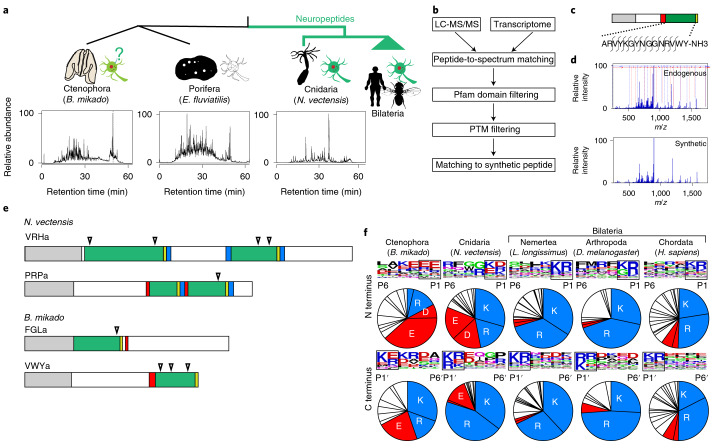
Fig. 1. Mass spectrometry-based identification of short amidated peptides in basal metazoans.
a, Phylogenic relationships of animals related to this study and known peptidergic systems are shown at the top. Total ion chromatograms of endogenous peptide fractions of basal metazoans N. vectensis (Cnidaria), E. fluviatilis (Porifera) and B. mikado (Ctenophora) are shown below. b, Data processing workflow of mass spectrometry-based neuropeptide identification. c, Schematic representation of the primary structure of the B. mikado VWYamide peptide. The positions of fragmentations observed in the fragment spectrum are indicated. d, Fragment spectra of endogenous and synthetic VWYamide. e, Schematic representation of neuropeptide precursors of FGLa, VWYa (B. mikado), PRPa and VRHa (N. vectensis). Grey, red, green and yellow boxes indicate signal peptide, cleavage site, mature neuropeptide and glycine as amide donor, respectively. Triangles denote the putative cleavage sites of neprilysin endopeptidase. No neuropeptide was detected from E. fluviatilis. f, Sequence logo map of N-terminal and C-terminal flanking regions and AA compositions of the cleavage sites (boxed) of neuropeptides in Metazoa. Basic (K and R) and acidic (D and E) AAs are shown in blue and red, respectively. Illustrations of fly, sea anemone and sponge are from phylopic.org.