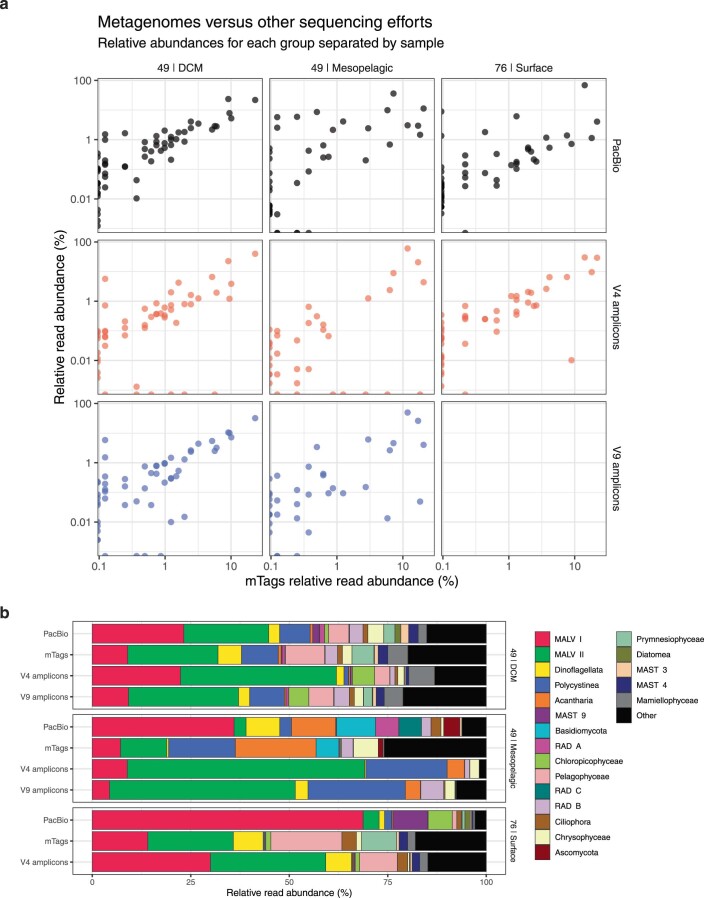
Extended Data Fig. 2. Metagenomes versus other sequencing efforts.
Comparison of the eukaryotic communities retrieved by PacBio and Illumina sequencing (V4, V9, and 18 S reads retrieved from metagenomic data) of three marine samples (See Extended Data Fig. 1). (a) Comparison of mTags (which should represent a snapshot of the community unbiased by PCR) with the other datasets. Groups explaining the majority of reads are detected at comparable abundances. Points at the margins represent taxa that are found in one dataset but not in the other; the line of dots along the y-axis represent groups not present in mTags, but present in other datasets; and along the x axis we see groups that are present in mTags but not in the other datasets. For instance in the 49|DCM panels, there are some groups recovered by mTags that V4/V9 amplicons cannot detect (blue and red points at the bottom). Groups detected by PacBio in 49|DCM but missed by V9 include: MAST-25, MOCH-1, Marine-Opisthokonts; whereas groups missed by V4 include: kinetoplastids, discoseans, diplonemids, pyrmnesiophytes, Marine-Opisthokonts, and Basal-Fungi. Fewer black points (PacBio) at the bottom of the panels, indicates that PacBio is detecting groups that are missed by metabarcoding with V4/V9 sequencing. (b) Overall comparison of the relative abundances at the group level (excluding Charophyta, Metazoa and Nucleomorphs). The primer pair used for long-read sequencing seem to preferentially amplify MALV-I, but the overall community structure that PacBio is retrieving is reasonable with the other sequencing approaches.