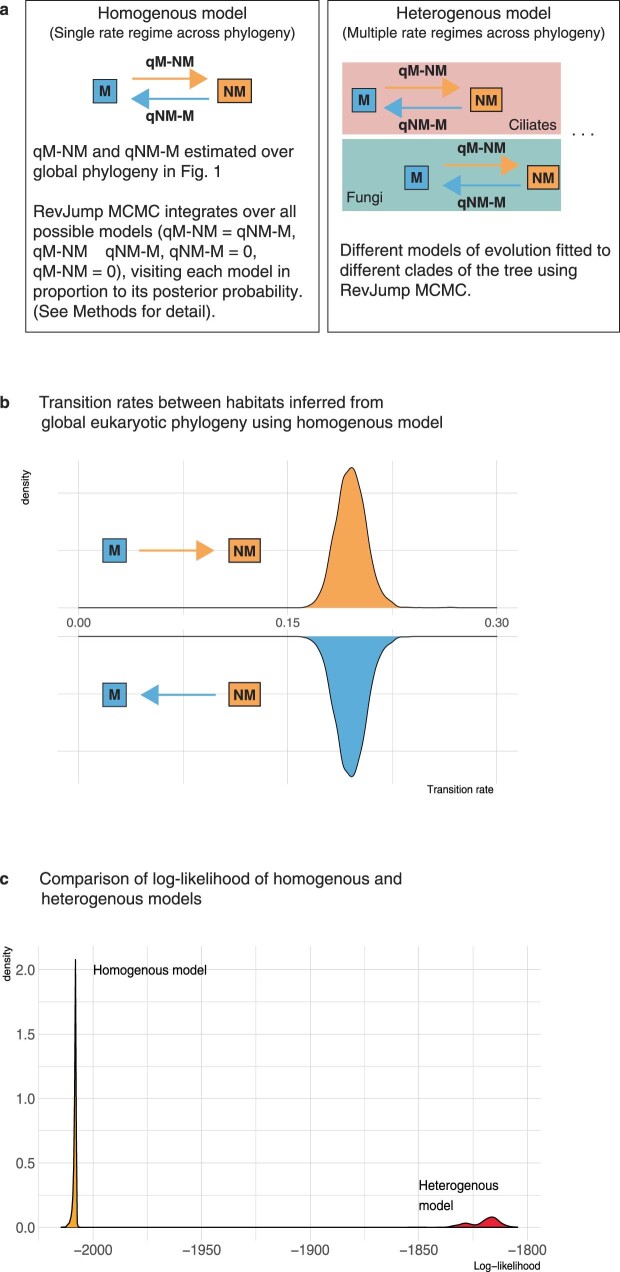
Extended Data Fig. 5. Testing models of habitat transitions on global eukaryotic phylogeny.
(a) A graphical representation of the homogenous and heterogeneous models tested. The homogenous model involves a single rate regime over the tree (that is qM−NM and qNM−M have constant values). No restrictions are placed on the parameters; so qM−NM and qNM−Mare allowed to be equal or unequal to each other. The heterogeneous model estimates a separate qM−NM and qNM−M for every major eukaryotic lineage (defined in this paper as rank 4 in the PR2-transitions database, for example Ciliates, Dinoflagellates, Fungi, etc.) that had at least 50 taxa and contained both marine and non-marine taxa. (b) Instantaneous transition rates from marine to non-marine habitats (qM−NM) and vice versa (qNM−M) when using a homogenous model over the global eukaryotic phylogeny (in Fig. 1). An equal rates model formulation (qM−NM = qNM−M) was found to have a higher posterior probability and was therefore sampled more frequently (100% of the time) by the reversible-jump Markov chain. (c) Comparison of the posterior probability of log-likelihoods when using the simple, homogenous model and the heterogeneous model. The plot shows that the heterogeneous model had a much better fit, indicating that rates of habitat evolution vary strongly across the eukaryotic tree of life.