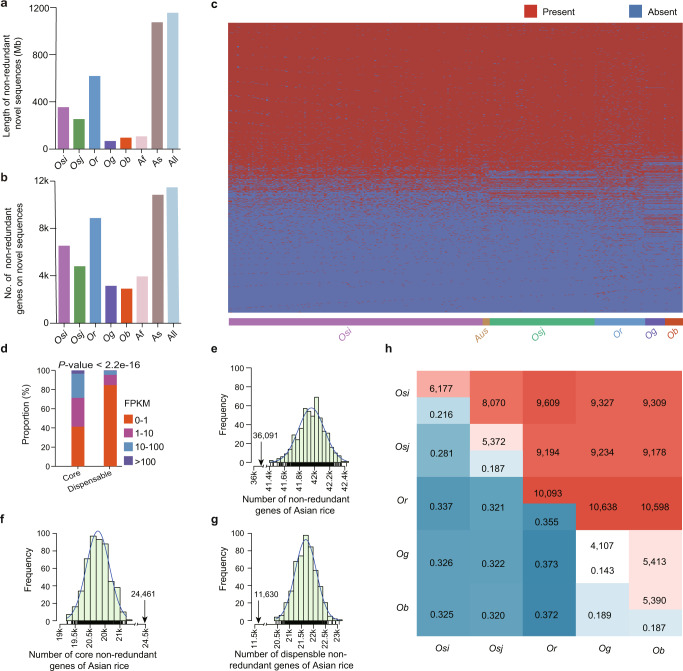
Fig. 2. Super pan-genome of 251 wild and cultivated Asian and African Oryza accessions.
a Total length of non-redundant novel sequences detected from the super pan-genome. Non-redundant novel sequences mean sequences that were absent in the Nipponbare reference genome and do not show redundancy across genomes. b Number of non-redundant genes in the non-redundant novel sequences. c The Landscape of PAVs for non-redundant genes across the 251 accessions. Each row indicates a non-redundant gene and each column indicates an accession. If the member of the non-redundant gene was present in an accession, it was colored in red; otherwise, it was colored in blue. Non-redundant genes were sorted by their occurrence. d Gene expression landscape of core and dispensable non-redundant genes. A Wilcoxon test was applied to the FPKM values. e–g Bootstrapping of all (e), core (f), and dispensable genes (g) in 11 Os and 10 Or. 11 Os and 10 Or were randomly selected 500 times from 230 Asian rice. The black arrows indicate the numbers of all, core and dispensable genes in 21 African rice (11 Og and 10 Ob), respectively. Non-redundant genes present in ≥ 95% accessions are defined as core non-redundant genes, and the rest of non-redundant genes are dispensable. h The average ratio (numbers) of different non-redundant genes when comparing two accessions. Os, Osi, Osj, Or, Og, Ob, Af, As and All respectively refer to O. sativa, O. sativa indica, O. sativa japonica, O. rufipogon, O. glaberrima, O. barthii, African rice, Asian rice and the 251 accessions.