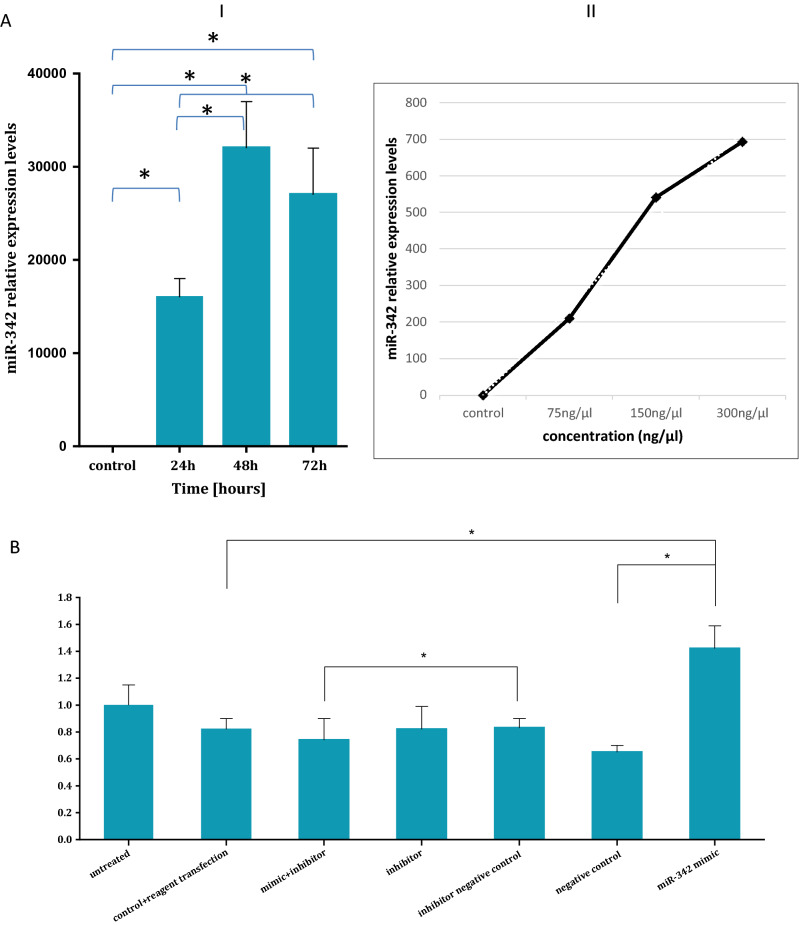
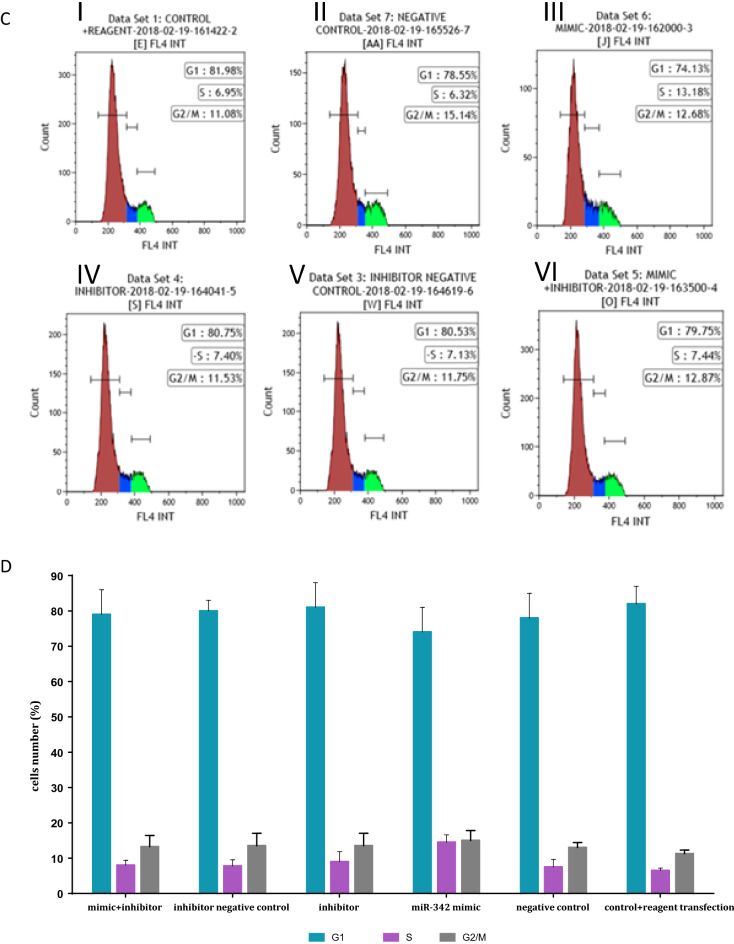
Figure 7.
miR342 provides fibroblasts cells with a survival advantage. (A) Transfection of pHFF cells with miR342. Left panel (I): Q-RT-PCR analysis of the relative expression levels of miR-342 after its mimic transfection. The standard Taqman MicroRNA Assays was followed to examine performance of RNU6B (snRNA) endogenous control candidates served as an internal reference. The data summarizes three experiments and expressed as mean ± SD for three experiments. (II) Right Panel: Q- RT-PCR analysis of the relative expression levels of miR342 mimic in the pHFF cells after treatment with various concentrations of that miR-342 mimic (75 ng/μl, 150 ng/μl, 300 ng/μl). * indicates statistical significance. (B). MiR-342 overexpression increases cellular proliferation. 96 h after 150 ng/ LmiR mimic 342 and its various controls transfection, we observed a statistically significant increase in the growth of miR-342 overexpressing cells in comparison to cells treated by negative control, untreated fibroblast, or fibroblast + reagent transfection (p < 0.05) By using the SRC assay. The data summarizes three independent experiments and expressed as mean ± SD. (C) The cell cycle status of pHFF cells in response to the ectopic expression of miR342. The DNA content of the cells was measured by flow cytometry. Cells were processed by standard methods using propidium iodide staining of DNA. (I) pHFF cell-cycle profile of cells transfected with transfection reagent. (II) pHFF cell-cycle profile of cells transfected with negative control. (III) pHFF cell-cycle profile of cells transfected with miR-342 mimic (IV) pHFF cell-cycle profile of cells transfected with miR-342 inhibitor (V) pHFF cell-cycle profile of cells transfected with inhibitor negative control (VI) pHFF cell-cycle profile of cells transfected with miR-342 mimic + miR342 inhibitor. (D) Quantification of 7C. Graph displaying differences in cell cycle phases, as determined by flow cytometry analysis, between normal and miR-342 expressing pHFF cells. The data summarizes three independent experiments and expressed as mean ± SD.