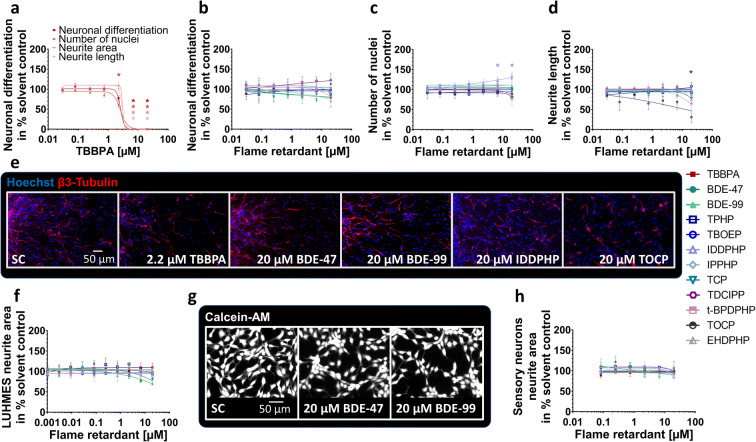
Fig. 4.
Neuronal differentiation and morphology (NPC3, NPC4, UKN4, UKN5) in the presence and absence of FRs. Spheres were plated onto poly-D-lysine/laminin-coated 96-well plates in the presence and absence of FRs. Differentiation into neurons (a, b) was determined automatically by using a convolutional neural network (CNN) running on Keras implemented in Python 3. The number of all β(III)tubulin-positive cells (red) in percent of Hoechst positive nuclei (blue) in the migration area after 120 h of differentiation was calculated (c, e). Morphology (d) was determined automatically by using the software Omnisphero. LUHMES cells and hiPSC derived sensory neurons were treated for 24 h in presence or absence of FRs and stained with Calcein-AM and H-33342 (g, LUHMES cells). An automated algorithm calculates the neurite area via subtraction of a calculated soma area from all calcein positive pixels (f, h). Data are represented as means ± SEM (except BDE-99 NPC3, n=2, means ± SD). Statistical significance was calculated using one-way ANOVA followed by Bonferroni’s post hoc tests (p < 0.05 was considered significant)