Fig. 1.
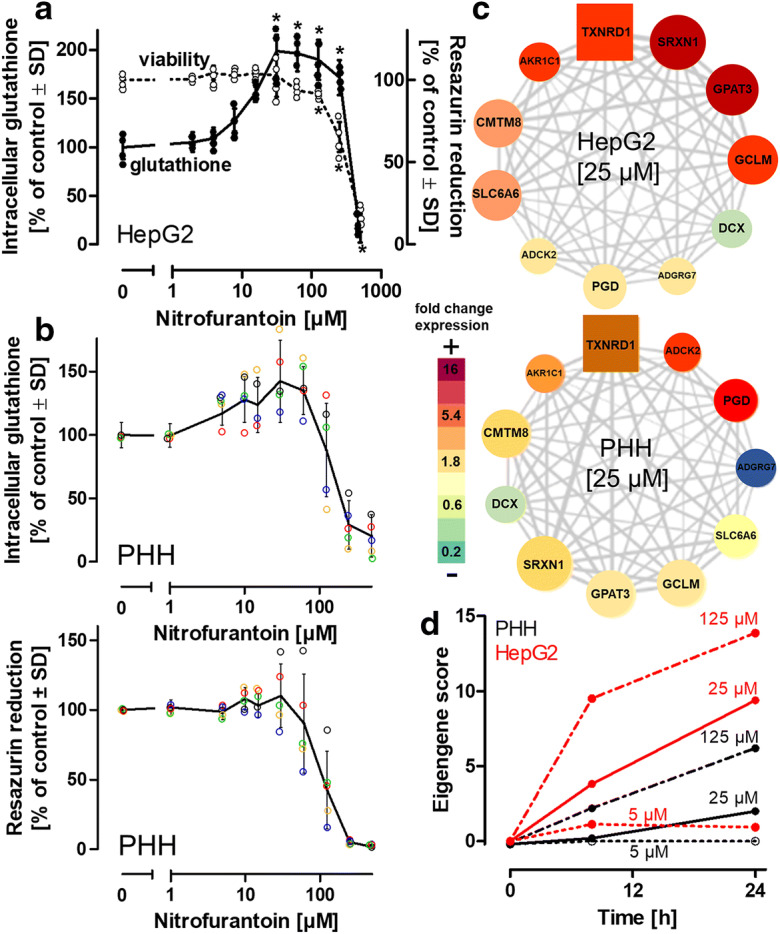
Elevation of glutathione by nitrofurantoin. a HepG2 (300,000 cells/cm2) were treated with indicated concentrations of nitrofurantoin (NFT). Intracellular levels of glutathione (reduced + oxidized) were detected after 48 h. Viability was assessed by measuring the reduction of resazurin. b Primary human hepatocytes (PHHs) from 5 donors were seeded at a density of 150,000 cells/cm2 and treated with NFT for 48 h. The colored circles represent values obtained with cells from 5 individual donors. The black solid lines represent the means of values from all five donors. Intracellular levels of glutathione (reduced + oxidized) were detected after 48 h. Viability was assessed by measuring the reduction of resazurin. c Gene co-expression analysis. Weighted gene co-expression network analysis was applied to the transcriptomic profiling of HepG2 exposed with NFT, utilizing the TXG-MAPr platform, allowing clustering of gene sets into functional modules. Significant changes in gene expression were identified for the module “oxidative stress” (module 144). The square node represents the gene with the highest correlation relative to the response of the entire module (hub gene). The size of the round nodes indicates the correlation of gene expression with the response of the entire module. The color code of the heatmap indicates up- or downregulation of the individual genes relative to their expression in untreated cells. d Eigengene scores indicating the sum of z-scores from the log2 fold changes of the individual genes of the module “oxidative stress” calculated in HepG2 or PHHs exposed for 8 h to NFT. Differences (in a; n = 5) were tested for significance by one-way ANOVA, followed by Bonferroni’s post hoc test, *p < 0.05 for comparison of treatments with the respective untreated controls. In (b), means ± SD are indicated by the solid line. Individual values obtained with cells from 4 different donors are indicated by colored dots. PGD 6-phosphogluconate dehydrogenase, SLC6A6 solute carrier family 6 member 7, CMTM8 CKLF-like MARVEL transmembrane domain containing 8, ADCK2 AarF domain containing kinase 2, ADGRG7 adhesion G protein-coupled receptor G7, DCX doublecortin, GCLM glutamate cysteine ligase modifier subunit, AKR1C1 Aldo-keto reductase family 1 member, TXNRD1 thioredoxin reductase 1, SRXN1 sulfiredoxin 1, GPAT3 glycerol-3-phosphate acetyltransferase 3
